Advanced Academic Degrees (M.Sc. and Ph.D.) in Medical Sciences
The faculty offers training in the medical sciences for students seeking to become researchers in academia or trained professional in industry and healthcare professions. The program includes frontal instruction, seminars, and research projects in the biological and medical sciences, laboratory science and clinical fields.
Eligible students are those who completed a bachelor degree (three or four year) in the life sciences, exact sciences or engineering. The applicants need to hold a minimum percentile grade point average of 85 in their undergraduate studies. The applicant is expected to find a prospective advisor from the faculty prior to registration.
Regardless of the undergraduate major, applicants must have an outstanding academic record. Relevant research experience is desirable. Students who were unable to obtain research experience are encouraged to obtain such experience prior to admission.
The graduate admissions committee reviews the application package (recommendation letters, transcripts, and resume) and interviews the prospective student to determine admission decisions.
The tabs below will guide candidates through the admissions process.
APPLICATIONApplications for the M.Sc. program are received with a specific deadline for the fall semester of the following year.
Before applying, the applicant must first identify a faculty member that will agree to serve as a thesis advisor. Find below a list of faculty members who are currently seeking M.Sc. students. To apply, please contact Ms. Shiri Shporen at the faculty of medicine graduate admissions office (04-829-5374).TUITIONTuition fees are determined at the time of admission.
Tuition fees for a magister’s degree are determined based on the study requirements given to the student at the time of admission. The thesis equals 20 credits, a final project equals 12 credits, and course load will be determined based on the academic background of the student. The tuition is determined for the entire degree, where 100% tuition means one year’s tuition. Thus, the tuition for a degree is usually 200-300%. If the student is not seeking a degree, the tuition will be 50% per semester. Upon admission, two forms (Bank form & Declaration form) need to be filled out and submitted to the Tuition Fees Departments.
Students who receive scholarships (most students at the faculty of medicine) are exempt from tuition and receive a monthly stipend (see FINANCIAL SUPPORT below for eligibility).
For details please either e-mail Ms. Shiri Shporen or call at 04-829-5374.
FINANCIAL SUPPORTScholarships and housing
For more information, see the Technion Graduate School website.
Ph.D. Degree in Medical Sciences
Only applicants holding M.Sc. or M.D. degrees and who excelled in their studies (percentile grade point average of 88 or above) will be accepted. The acceptance of applicants with international degrees will be pending approval of the Graduate School of Technion. The decision of the departmental Graduate Studies Committee will be based on grades, curriculum vitae, professional achievements and letters of recommendation. The applicant is expected to find a prospective advisor from the Faculty staff, prior to registration. Forms of “Agreement to Serve as Advisor” are available at the departmental Office for Graduate Studies
APPLICATIONApplications for the Ph.D. program are received year-round.
Before applying, the applicant must first identify a faculty member that will agree to serve as a thesis advisor. Find below a list of faculty members who are currently seeking Ph.D. students. To apply, please contact Ms. Shiri Shporen at the faculty of medicine graduate admissions office (04-829-5374).FINANCIAL SUPPORTScholarships and housing
For more information, see the Graduate School website.
Find an Advisor
The applicant is expected to find a prospective advisor prior to registration. Prospective students are encouraged to identify potential faculty members and contact them directly.
If you would like to receive general guidance, you can contact Prof. Oded Lewinson , the graduate program coordinator. For application process questions and an Agreement to Serve as Advisor form, contact Ms. Shiri Shporen, the graduate program administrator.
Genetics and Developmental Biology
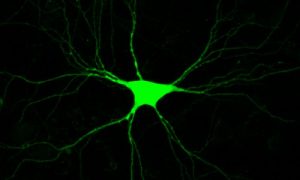
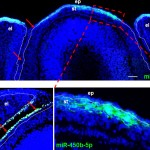
The Genetics of Vision and Hearing Research Laboratory of Associate Professor Tamar Ben-Yosef.  My research focuses mainly on inherited retinal degenerations (IRD), a heterogeneous group of diseases, which cause visual loss due to the premature death of photoreceptors in the retina. My research combines genetic and functional studies. I recruit IRD patients and their families, and perform various genetic analyses to identify the genetic cause for disease in each family. Identified mutations are studied at the population level, to identify common founder mutations in certain Israeli ethnic groups. Identified genes are further studied to characterize their retinal expression pattern and function.
My research focuses mainly on inherited retinal degenerations (IRD), a heterogeneous group of diseases, which cause visual loss due to the premature death of photoreceptors in the retina. My research combines genetic and functional studies. I recruit IRD patients and their families, and perform various genetic analyses to identify the genetic cause for disease in each family. Identified mutations are studied at the population level, to identify common founder mutations in certain Israeli ethnic groups. Identified genes are further studied to characterize their retinal expression pattern and function.
 The Laboratory of Tissue Development of Associate Professor Peleg Hasson. The extracellular matrix and the connective tissues that secrete it play cardinal instructive roles, regulating numerous processes in development and regeneration, health and disease. Although many of the matrix’s components have been identified, understanding of how its building blocks are laid and organized and how these properties affect the underlying cells and tissues are still largely unclear. Using mouse genetics and cell-culture based assays, our lab focuses on two major processes which the matrix and connective tissues contribute to: the development and regeneration of the muscles and tendons as well as the development, maturation and remodeling of the blood vasculature.
The Laboratory of Tissue Development of Associate Professor Peleg Hasson. The extracellular matrix and the connective tissues that secrete it play cardinal instructive roles, regulating numerous processes in development and regeneration, health and disease. Although many of the matrix’s components have been identified, understanding of how its building blocks are laid and organized and how these properties affect the underlying cells and tissues are still largely unclear. Using mouse genetics and cell-culture based assays, our lab focuses on two major processes which the matrix and connective tissues contribute to: the development and regeneration of the muscles and tendons as well as the development, maturation and remodeling of the blood vasculature.
The Evolutionary Process of Mutation & Natural Selection Research Laboratory of Associate Professor Ruth Hershberg. Many medically relevant phenomena are in fact evolutionary processes. These include cancer initiation and progression, pathogen emergence and adaptation and the emergence and spread of antibiotic resistance within bacterial populations. In our lab we aim to use evolutionary insights, tools and perspectives, together with cutting edge molecular, genomic and bioinformatics techniques to study such medically relevant evolutionary processes. More specific areas of research include (but are not limited to): (1) What are the antibiotic-independent fitness effects of antibiotic resistance mutations and how do these affect the spread of resistance within natural bacterial populations? (2) Using evolutionary insights and tools to identify cancer driving genes (3) Pathogen evolution via gene loss.
 The Developmental Genetics Research Laboratory of Associate Professor Adi Salzberg. One of the fascinating questions in developmental biology is how different cells, which develop within a given tissue, acquire different properties that allow them to work together as a functional unit. We are using the proprioceptive organs in the peripheral nervous system (PNS) of Drosophila(also called chordotonal organs, ChOs) as a model system for studying cell fate diversification. The fly ChOs, sensory organs that detect motion and position, are composed of linear arrays of lineage-related cells with distinct identities and properties that allow them to work together as a functional organ. Very little is known about the mechanisms regulating the specification of the various ChO cell identities. Through our work on the deigene we have identified distinct regulatory elements that direct gene expression to specific ChO cell types. We have used these elements to construct a unique transgenic fly strain in which different ChO cell types are labeled with different fluorescent markers. This strain is currently being used to conduct a large-scale, RNAi-based, genetic screen for novel determinants of cell fate specification and morphogenesis in the ChO lineage.
The Developmental Genetics Research Laboratory of Associate Professor Adi Salzberg. One of the fascinating questions in developmental biology is how different cells, which develop within a given tissue, acquire different properties that allow them to work together as a functional unit. We are using the proprioceptive organs in the peripheral nervous system (PNS) of Drosophila(also called chordotonal organs, ChOs) as a model system for studying cell fate diversification. The fly ChOs, sensory organs that detect motion and position, are composed of linear arrays of lineage-related cells with distinct identities and properties that allow them to work together as a functional organ. Very little is known about the mechanisms regulating the specification of the various ChO cell identities. Through our work on the deigene we have identified distinct regulatory elements that direct gene expression to specific ChO cell types. We have used these elements to construct a unique transgenic fly strain in which different ChO cell types are labeled with different fluorescent markers. This strain is currently being used to conduct a large-scale, RNAi-based, genetic screen for novel determinants of cell fate specification and morphogenesis in the ChO lineage.
The Synaptic Tenacity and Maintenance Laboratory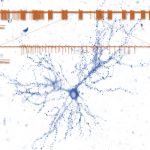 of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
___________________________________________________________________________________________________________________
 The Epithelial Stem Cells and Pathophysiology Research Laboratory of Assistant Professor Ruby Shalom-Feuerstein. Our lab explores the molecular events that underlie the morphogenesis, stem cell homeostasis, and pathophysiology of the skin and the cornea. We employ complementary approaches, some of which were uniquely developed in our lab, to investigate these biological processes and develop novel therapies. Among these technologies are induced pluripotent stem cell and adult stem cell cultures, lineage tracing experiments and genetic mouse models, molecular biology, and genome wide analysis.
The Epithelial Stem Cells and Pathophysiology Research Laboratory of Assistant Professor Ruby Shalom-Feuerstein. Our lab explores the molecular events that underlie the morphogenesis, stem cell homeostasis, and pathophysiology of the skin and the cornea. We employ complementary approaches, some of which were uniquely developed in our lab, to investigate these biological processes and develop novel therapies. Among these technologies are induced pluripotent stem cell and adult stem cell cultures, lineage tracing experiments and genetic mouse models, molecular biology, and genome wide analysis.
The Laboratory of Organogenesis and Developmental Biology of Associate Professor Tom Schultheiss.  The fields of Regenerative Medicine and Tissue Engineering, which aim to generate tissues in the laboratory in order to replace and repair damaged organs, hold great promise for treating many congenital and chronic diseases. In order to generate tissues and organs in the lab, it essential to understand how these tissues are normally built in the developing embryo. The Schultheiss laboratory studies how organs are built in the embryo, from the first steps when embryonic cells adopt tissue-specific identities, through the later stages when the functional organ is assembled from its various cellular components. Our current work focuses on the formation of the kidney, gonads, and hematopoietic system, and we use the avian embryo (chicken and quail) as our main experimental system. We also have a strong interest in understanding basic embryological processes, such as embryonic patterning, cell differentiation, migration, and morphogenesis.
The fields of Regenerative Medicine and Tissue Engineering, which aim to generate tissues in the laboratory in order to replace and repair damaged organs, hold great promise for treating many congenital and chronic diseases. In order to generate tissues and organs in the lab, it essential to understand how these tissues are normally built in the developing embryo. The Schultheiss laboratory studies how organs are built in the embryo, from the first steps when embryonic cells adopt tissue-specific identities, through the later stages when the functional organ is assembled from its various cellular components. Our current work focuses on the formation of the kidney, gonads, and hematopoietic system, and we use the avian embryo (chicken and quail) as our main experimental system. We also have a strong interest in understanding basic embryological processes, such as embryonic patterning, cell differentiation, migration, and morphogenesis.
 Laboratory for Mechanobiology of the Cell of Assistant Professor Haguy Wolfenson. Cells are exposed to a variety of signals that control their fate. Whereas biomedical studies are mostly focused on biochemical signals, the mechanical features of the microenvironment around cells are as much important in regulating cellular functions such as migration, survival, growth, and differentiation. The studies in the lab are focused on the mechanisms by which cells sense and respond to mechanical features of their environment such as rigidity, topography, or ligand density. We use a combination of advanced microscopy techniques with biophysical measurements to decipher the mechanisms by which extracellular mechanical signals are transmitted into biochemical signals inside the cells and how those in turn affect cellular behavior. We also study how these processes are altered in cancer with the premise that proper mechanosensing is malfunctioning in cancer cells.
Laboratory for Mechanobiology of the Cell of Assistant Professor Haguy Wolfenson. Cells are exposed to a variety of signals that control their fate. Whereas biomedical studies are mostly focused on biochemical signals, the mechanical features of the microenvironment around cells are as much important in regulating cellular functions such as migration, survival, growth, and differentiation. The studies in the lab are focused on the mechanisms by which cells sense and respond to mechanical features of their environment such as rigidity, topography, or ligand density. We use a combination of advanced microscopy techniques with biophysical measurements to decipher the mechanisms by which extracellular mechanical signals are transmitted into biochemical signals inside the cells and how those in turn affect cellular behavior. We also study how these processes are altered in cancer with the premise that proper mechanosensing is malfunctioning in cancer cells.
The Laboratory of Gene Regulation and Phenotypic Evolution of Assistant Professor Ella Preger-Ben Noon. Our lab studies the molecular mechanisms underlying enhancer function and  evolution. We investigate how mutations in these regulatory DNA sequences lead to the evolution of animal body form. To address these questions, we combine the powerful genetic toolkit of the fruit fly Drosophila with cutting-edge techniques of cell type-specific genomics, single-molecule live imaging and genome editing. Our approach allows us to untangle the precise molecular mechanisms that underlie phenotypic changes between closely related species and to uncover new models of gene regulation.
evolution. We investigate how mutations in these regulatory DNA sequences lead to the evolution of animal body form. To address these questions, we combine the powerful genetic toolkit of the fruit fly Drosophila with cutting-edge techniques of cell type-specific genomics, single-molecule live imaging and genome editing. Our approach allows us to untangle the precise molecular mechanisms that underlie phenotypic changes between closely related species and to uncover new models of gene regulation.
Cell Biology and Cancer Science
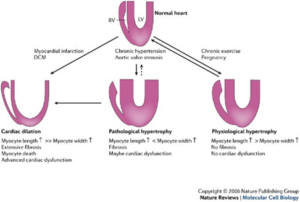 The role of ATF3/JDP2 bZIP proteins in heart pathophysiology laboratory of Prof. Ami Aronheim. Hypertrophic growth of the cardiac muscle is an adaptive response that occurs in physiological and pathological conditions. However, whereas physiological hypertrophy is beneficial for heart function, sustained pathological hypertrophy leads to cardiac maladaptive remodeling processes resulting in permanent contractile dysfunction and heart failure. Currently, the incidence of heart failure is on the rise and treatment options are limited. Cardiac remodeling is regulated by multiple G-protein coupled receptors that converge to a limited number of transcription factors. Thus, targeting key transcriptional regulators emerges as a promising strategy for the development of new therapeutic approaches. Over the last decade, we have studied the role of the Activating Transcription Factor 3 (ATF3), a basic leucine zipper protein (bZIP), in promoting maladaptive cardiac remodeling processes in response to pressure overload. ATF3 is most closely homologous to the c-Jun Dimerization Protein 2 (JDP2), a bZIP gene that was initially isolated in our lab. ATF3 and JDP2 are primarily transcription inhibitors. Our results show that hearts derived from mice lacking both ATF3 and JDP2 are enlarged, exhibit an improved contractile function, however, with no apparent increase in either hypertrophic, fibrosis or inflammatory markers. In addition, these mice are protected from pressure overload-induced maladaptive cardiac remodeling. We aim to study the molecular mechanisms leading to protection of the heart from maladaptive cardiac remodeling processes. Ultimately, this study will provide new insights towards the development of novel strategies for the treatment of cardiac dysfunction and heart failure.
The role of ATF3/JDP2 bZIP proteins in heart pathophysiology laboratory of Prof. Ami Aronheim. Hypertrophic growth of the cardiac muscle is an adaptive response that occurs in physiological and pathological conditions. However, whereas physiological hypertrophy is beneficial for heart function, sustained pathological hypertrophy leads to cardiac maladaptive remodeling processes resulting in permanent contractile dysfunction and heart failure. Currently, the incidence of heart failure is on the rise and treatment options are limited. Cardiac remodeling is regulated by multiple G-protein coupled receptors that converge to a limited number of transcription factors. Thus, targeting key transcriptional regulators emerges as a promising strategy for the development of new therapeutic approaches. Over the last decade, we have studied the role of the Activating Transcription Factor 3 (ATF3), a basic leucine zipper protein (bZIP), in promoting maladaptive cardiac remodeling processes in response to pressure overload. ATF3 is most closely homologous to the c-Jun Dimerization Protein 2 (JDP2), a bZIP gene that was initially isolated in our lab. ATF3 and JDP2 are primarily transcription inhibitors. Our results show that hearts derived from mice lacking both ATF3 and JDP2 are enlarged, exhibit an improved contractile function, however, with no apparent increase in either hypertrophic, fibrosis or inflammatory markers. In addition, these mice are protected from pressure overload-induced maladaptive cardiac remodeling. We aim to study the molecular mechanisms leading to protection of the heart from maladaptive cardiac remodeling processes. Ultimately, this study will provide new insights towards the development of novel strategies for the treatment of cardiac dysfunction and heart failure.
___________________________________________________________________________________________________________________
 Aaron Ciechanover, MD, DSc Distinguished Professor Laureate, the 2004 Nobel Prize in Chemistry The ubiquitin proteolytic system, pathogenesis of diseases,
Aaron Ciechanover, MD, DSc Distinguished Professor Laureate, the 2004 Nobel Prize in Chemistry The ubiquitin proteolytic system, pathogenesis of diseases,
and development of therapeutic modalities.
Our group is part of the Rappaport – Technion Integrated Cancer Center – R-TICC and we work in tight collaboration with many of its researchers. Our studies focus on the role of the ubiquitin system (that was discovered in our laboratory and was culminated with the awarding of the 2004 Nobel Prize in Chemistry) in basic mechanisms of diseases, cancer and neurodegeneration in particular. Our research is an elegant example of how curiosity-driven basic research leads – by different groups – to its translation to efficient drugs to combat severe human diseases – including malignancies and neurodegeneration.
One area of studies is the mechanism by of activation of NF-κB, a major transcription factor that is upregulated in many tumors. We identified a surprising side to this known story when we showed that overexpression of its p50 subunit, which is typically generated in stoichiometric amounts to the other partner – p65, displays a strong tumor suppressive effect that we now study.
Another area of studies involves the proteasome, which is the catalytic arm of the ubiquitin system. Here we showed that by simple metabolic “trick” we can lock it in the nucleus, preventing it from supplying the cell with amino acids derived from degradation of intracellular proteins, and thus resulting in cell death. It appears that cancer cells are more sensitive to this lock, as their metabolic needs are much larger than those of healthy cells.
In an independent line of research, we are studying the involvement of the ubiquitin system in neurodegenerative disorders – the hallmark of which is accumulation in the brain of abnormal/mutated and typically aggregated proteins that are not recognized by the ubiquitin system. Out of several diseases, we decided to study Huntington’s disease where several good experimental tools – probes, cells, and animal models – are available. Huntington’s disease (HD) is a progressive incurable neurodegenerative disorder characterized by motor and neuropsychiatric symptoms
Cancer Metabolism Laboratory of Professor Eyal Gottlieb. We utilize metabolomics approaches to identify metabolic alterations and to target metabolic essential pathways of cancer cells.
The Computational Cancer Genomics lab of Dr. Yizhak focuses on computational challenges in cancer research. As opposed to traditional “wet” labs, much of the biological and medical  research today is done computationally in what we call “dry” labs. Such research is enabled by large amount of clinical and molecular data collected around the world by the scientific community.
research today is done computationally in what we call “dry” labs. Such research is enabled by large amount of clinical and molecular data collected around the world by the scientific community.
Our lab addresses emerging challenges in computational cancer genomics that require the development and application of innovative computational methods and tools for cancer data analysis. We work closely with clinicians and experimentalists in Israel and around the world to turn data into knowledge and advance cancer treatment and prevention.
 Genetic Networks, Tissue Homeostasis and Cancer Laboratory of Associate Professor Amir Orian (Oryan). My laboratory focuses on understanding fundamental mechanisms and genes of cancer-related genetic networks. These networks are critical for maintenance of the differentiated identity and deregulation of such networks is associated with cancer. We focus on: 1. “Identity network”;, genes that are required to maintain the identity of differentiated cells and serve as a “barrier to tumorigenesis.” 2. NOA-gene network; Non-Oncogene Addiction genes that a essential for cancer cells to cope with the oncogenic stress, are vital for maintaining the tumorous phenotype, but are less critical to un-transformed cells. Thus, they are have the potential to be molecular targets for molecular cancer therapy. Our current projects include: 1. Characterizing the role of SUMO-Targeted-Ubiquitin Ligase proteins in Drosophila development and human cancer; 2. Identifying genes involved in innate immune responses to distinct pathogens; and 3. Delineating the genes that regulate adult gut homeostasis at the genetic and genomic level, with a focus on genes that maintain the balance between progenitor and differentiated cells. Towards these aims we employ advanced genetic and genomic tools to study transcriptional networks using Drosophila genetics and genomics, mammalian cells, mouse-derived intestinal organoids and patient-derived specimens.
Genetic Networks, Tissue Homeostasis and Cancer Laboratory of Associate Professor Amir Orian (Oryan). My laboratory focuses on understanding fundamental mechanisms and genes of cancer-related genetic networks. These networks are critical for maintenance of the differentiated identity and deregulation of such networks is associated with cancer. We focus on: 1. “Identity network”;, genes that are required to maintain the identity of differentiated cells and serve as a “barrier to tumorigenesis.” 2. NOA-gene network; Non-Oncogene Addiction genes that a essential for cancer cells to cope with the oncogenic stress, are vital for maintaining the tumorous phenotype, but are less critical to un-transformed cells. Thus, they are have the potential to be molecular targets for molecular cancer therapy. Our current projects include: 1. Characterizing the role of SUMO-Targeted-Ubiquitin Ligase proteins in Drosophila development and human cancer; 2. Identifying genes involved in innate immune responses to distinct pathogens; and 3. Delineating the genes that regulate adult gut homeostasis at the genetic and genomic level, with a focus on genes that maintain the balance between progenitor and differentiated cells. Towards these aims we employ advanced genetic and genomic tools to study transcriptional networks using Drosophila genetics and genomics, mammalian cells, mouse-derived intestinal organoids and patient-derived specimens.
The Tumor Biology Lab of Professor Vlodavskyfocuses on cell interaction with the extracellular matrix (ECM) and the tumor microenvironment. In the center of this research is an enzyme (heparanase) which degrades a major component (heparan sulfate) of the ECM and thereby releases numerous factors that promote tumor growth, metastasis and angiogenesis. Heparanase-inhibiting compounds and neutralizing antibodies are being developed to inhibit cancer progression and inflammation. Among the issues that are being investigated in the Lab are: 1)How dose heparanase activate macrophages and other cells of the immune system? 2) How does heparanase elicit chemoresistance? 3) Can heparanase inhibitors block tumor progression and chemoresistance driven by host heparanase? 4) What is the role of heparanase-2 in normal physiology? and 5) Do heparanase-1 and heparanse-2 play a role in gene transcription?
Biochemistry
 The Redox Biology Laboratory of Associate Professor Benhar Moran focuses on studying the mechanisms whereby redox modifications of cysteine residues regulate protein and cell function. Research projects in the lab use cutting-edge proteomic and biochemical tools to explore the roles of protein redox modifications (such as cysteine nitrosylation) in cellular communication, inflammation, and cancer.
The Redox Biology Laboratory of Associate Professor Benhar Moran focuses on studying the mechanisms whereby redox modifications of cysteine residues regulate protein and cell function. Research projects in the lab use cutting-edge proteomic and biochemical tools to explore the roles of protein redox modifications (such as cysteine nitrosylation) in cellular communication, inflammation, and cancer.
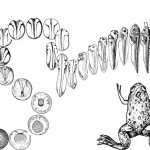 The Molecular Embryology Laboratory of Associate Professor Dale Frank. My research group studies the initial generation of the body axis occurring during embryonic development. Using amphibians as a model vertebrate system, we investigate the cross-talk between transcription factors and signaling pathways to determine how they induce neural and mesodermal cell fates along the body axis during early embryo development. Frogs, like people, are vertebrates, and the regulation of early embryonic development in all vertebrates (including mammals) is carried out by the same gene products regardless, of species. Thus, the frog (Xenopus laevis) is an excellent system to learn how all vertebrates make their body plan. One of the earliest and most dramatic events in early development is the formation of the body axes. As early embryos, we all start out as a round egg cell, which then rapidly divides into groups of cells. These early stem cells are totipotential and can develop into all cell types of the embryo. At a critical stage, cells commit to specific fates. The cells start migrating, and they elongate to make our typical body plan: the head – tail axis (anterior – posterior), the back – stomach axis (dorsal – ventral) and finally the left – right axis. Cells must know what they are doing along theses body axes. Not only must a cell decide its own identity, a decision to be muscle, blood or nerve for instance. Cells must also define their position along the axis. In the nervous system for example, a cell in the anterior-head makes a forebrain-derived neuron or an eye cell, whereas a nerve cell in the posterior-tail region makes a motor neuron. This location-dependence of cell fate is based on the capability of cells to monitor their position with respect to the developing embryonic axes. In my lab, we are working on the genetic and cellular interactions determining how progenitor like stem-like of the early mesoderm and nervous system acquire these different axial cell fates during early development.
The Molecular Embryology Laboratory of Associate Professor Dale Frank. My research group studies the initial generation of the body axis occurring during embryonic development. Using amphibians as a model vertebrate system, we investigate the cross-talk between transcription factors and signaling pathways to determine how they induce neural and mesodermal cell fates along the body axis during early embryo development. Frogs, like people, are vertebrates, and the regulation of early embryonic development in all vertebrates (including mammals) is carried out by the same gene products regardless, of species. Thus, the frog (Xenopus laevis) is an excellent system to learn how all vertebrates make their body plan. One of the earliest and most dramatic events in early development is the formation of the body axes. As early embryos, we all start out as a round egg cell, which then rapidly divides into groups of cells. These early stem cells are totipotential and can develop into all cell types of the embryo. At a critical stage, cells commit to specific fates. The cells start migrating, and they elongate to make our typical body plan: the head – tail axis (anterior – posterior), the back – stomach axis (dorsal – ventral) and finally the left – right axis. Cells must know what they are doing along theses body axes. Not only must a cell decide its own identity, a decision to be muscle, blood or nerve for instance. Cells must also define their position along the axis. In the nervous system for example, a cell in the anterior-head makes a forebrain-derived neuron or an eye cell, whereas a nerve cell in the posterior-tail region makes a motor neuron. This location-dependence of cell fate is based on the capability of cells to monitor their position with respect to the developing embryonic axes. In my lab, we are working on the genetic and cellular interactions determining how progenitor like stem-like of the early mesoderm and nervous system acquire these different axial cell fates during early development.
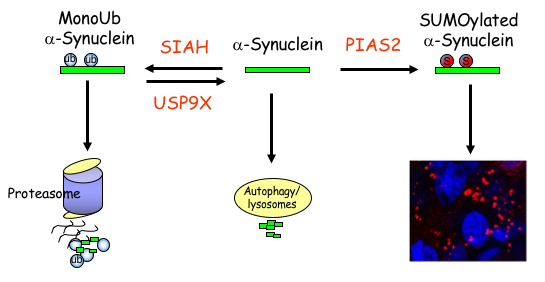 The Laboratory of Neurodegeneration of Associate Prof. Simone Engelender. Parkinson’s disease (PD) is a very common neurodegenerative disease and it is caused by the neuronal accumulation of a protein known as a-synuclein. Our goal is to investigate the mechanisms that lead to the accumulation of a-synuclein in the brain of PD patients. We found that monoubiquitination of a-synuclein leads to its degradation by the proteasome. Nevertheless, the degradation of monoubiquitinated a-synuclein is impaired in PD, leading to the accumulation and aggregation of a-synuclein. More recently, we found that additional post-translational modifications, such as SUMOylation, prevents the proteasomal degradation of a-synuclein and leads to even further accumulation and aggregation of a-synuclein into inclusions, which resemble to the inclusions found in the disease. In addition, we identified several proteins of the monoubiquitination and SUMOylation pathways that are dysfunctional in PD brains, further suggesting their role in a-synuclein homeostasis and PD. We are currently utilizing advanced biochemical and cell biology techniques as well as generating novel PD-related transgenic mice to further investigate the mechanisms that regulate a-synuclein degradation, accumulation and aggregation in PD. In an attempt to decrease a-synuclein levels in the disease, we are also generating compounds to specifically target the a-synuclein monoubiquitination and SUMOylation pathways.
The Laboratory of Neurodegeneration of Associate Prof. Simone Engelender. Parkinson’s disease (PD) is a very common neurodegenerative disease and it is caused by the neuronal accumulation of a protein known as a-synuclein. Our goal is to investigate the mechanisms that lead to the accumulation of a-synuclein in the brain of PD patients. We found that monoubiquitination of a-synuclein leads to its degradation by the proteasome. Nevertheless, the degradation of monoubiquitinated a-synuclein is impaired in PD, leading to the accumulation and aggregation of a-synuclein. More recently, we found that additional post-translational modifications, such as SUMOylation, prevents the proteasomal degradation of a-synuclein and leads to even further accumulation and aggregation of a-synuclein into inclusions, which resemble to the inclusions found in the disease. In addition, we identified several proteins of the monoubiquitination and SUMOylation pathways that are dysfunctional in PD brains, further suggesting their role in a-synuclein homeostasis and PD. We are currently utilizing advanced biochemical and cell biology techniques as well as generating novel PD-related transgenic mice to further investigate the mechanisms that regulate a-synuclein degradation, accumulation and aggregation in PD. In an attempt to decrease a-synuclein levels in the disease, we are also generating compounds to specifically target the a-synuclein monoubiquitination and SUMOylation pathways.
The Molecular Machines laboratory 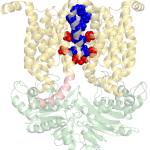 of Associate Professor Oded Lewinson. Our lab is interested in how molecular machines are built and how they work. We are especially interested in molecular machines that are important for cancer multidrug resistance and for bacterial pathogenesis. We harness this knowledge to identify new targets for drug development.
of Associate Professor Oded Lewinson. Our lab is interested in how molecular machines are built and how they work. We are especially interested in molecular machines that are important for cancer multidrug resistance and for bacterial pathogenesis. We harness this knowledge to identify new targets for drug development.
For example, in 2018 our lab received the NATO Prize for Peace and Security following our discovery that zinc can be used as a highly effective inhibitor of an essential virulence factor of Anthrax (shown here on the right).
Structure of the Bacillus Anthracis
virulence determinant MntBC and
its drug binding sites
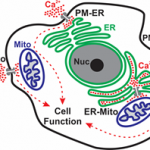 Calcium Signaling Laboratory of Senior Lecturer Raz Palty. Calcium ions are central and ubiquitous intracellular signaling entities. Changes in intracellular calcium levels control fundamental cellular events, from signaling the mitotic birth of a new cell to marking its end during apoptosis. To generate calcium signals, cells rely on an elaborate machinery of ion transport proteins that are strategically positioned in the plasma membrane (PM) and intracellular organelle membranes. The function of calcium transport proteins at these sites is critical for numerous physiological processes including synaptic transmission, muscle contraction, and the immune response. Consequently, several disease states arise due to dysfunction of calcium transport mechanisms including neurological disorders, cardiac disease and immunodeficiency. Our main focus in the Palty lab is to elucidate molecular mechanisms that regulate calcium signaling across the PM, endoplasmic reticulum (ER) and mitochondria with the long-term goal of understanding how calcium signals are generated at these sites under normal physiological conditions and in disease states.
Calcium Signaling Laboratory of Senior Lecturer Raz Palty. Calcium ions are central and ubiquitous intracellular signaling entities. Changes in intracellular calcium levels control fundamental cellular events, from signaling the mitotic birth of a new cell to marking its end during apoptosis. To generate calcium signals, cells rely on an elaborate machinery of ion transport proteins that are strategically positioned in the plasma membrane (PM) and intracellular organelle membranes. The function of calcium transport proteins at these sites is critical for numerous physiological processes including synaptic transmission, muscle contraction, and the immune response. Consequently, several disease states arise due to dysfunction of calcium transport mechanisms including neurological disorders, cardiac disease and immunodeficiency. Our main focus in the Palty lab is to elucidate molecular mechanisms that regulate calcium signaling across the PM, endoplasmic reticulum (ER) and mitochondria with the long-term goal of understanding how calcium signals are generated at these sites under normal physiological conditions and in disease states.
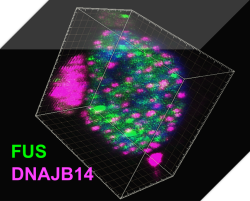 Protein homeostasis regulatory networks – A systems biology approach to protein homeostasis regulation in stress and neurodegenerative diseases.The lab of Associate Professor Reut Shalgi integrates molecular and computational biology towards the goal of understanding gene expression regulation, and protein homeostasis networks, in stressed human cells, and how they are impaired in neurodegenerative diseases. We mainly focus on Huntington’s disease, and ALS, and use cutting-edge technologies, including NGS transcriptomics and translatomics, protein-protein interaction network mapping, high throughput screening, and big data analysis, to understand the mechanisms of these diseases, and reveal how we can help the cells with their battle against them.
Protein homeostasis regulatory networks – A systems biology approach to protein homeostasis regulation in stress and neurodegenerative diseases.The lab of Associate Professor Reut Shalgi integrates molecular and computational biology towards the goal of understanding gene expression regulation, and protein homeostasis networks, in stressed human cells, and how they are impaired in neurodegenerative diseases. We mainly focus on Huntington’s disease, and ALS, and use cutting-edge technologies, including NGS transcriptomics and translatomics, protein-protein interaction network mapping, high throughput screening, and big data analysis, to understand the mechanisms of these diseases, and reveal how we can help the cells with their battle against them.
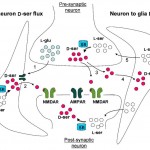 The Molecular Neuroscience Laboratory of Professor Herman Wolosker. Research in our laboratory focuses on understanding the roles of unconventional neurotransmitters, like the D-amino acids in the central nervous system. We are particularly interested in the regulation of N-Methyl- D-aspartate receptors (NMDARs) by D-serine, a D-enantiomer previously thought to be restricted to bacteria or lower invertebrates. D-Serine is now increasingly appreciated as a major physiologic ligand for NMDARs that mediates NMDAR synaptic responses and neurotoxicity both in vitro and in vivo. In addition to neurotransmission, NMDARs play a key role in neurodegeneration, with their excessive activation contributing to neuronal death in several neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis. Over the past few years, we elucidated the mechanisms of D-serine synthesis by the enzyme serine racemase and discovered novel pathways regulating D-serine production and release from neural cells. We also discovered that D-serine is the dominant NMDAR co-agonist mediating neurotoxicity, raising the possibility that drugs that curb D-serine synthesis or release might be useful in neurodegenerative diseases involving NMDARs over-stimulation. We use molecular biology, biochemical and cell biology techniques in tissues and cell culture models. We generate and keep new mouse genetic models of brain transporters and metabolic enzymes, and also apply neurobiological techniques both in vitro and in vivo.
The Molecular Neuroscience Laboratory of Professor Herman Wolosker. Research in our laboratory focuses on understanding the roles of unconventional neurotransmitters, like the D-amino acids in the central nervous system. We are particularly interested in the regulation of N-Methyl- D-aspartate receptors (NMDARs) by D-serine, a D-enantiomer previously thought to be restricted to bacteria or lower invertebrates. D-Serine is now increasingly appreciated as a major physiologic ligand for NMDARs that mediates NMDAR synaptic responses and neurotoxicity both in vitro and in vivo. In addition to neurotransmission, NMDARs play a key role in neurodegeneration, with their excessive activation contributing to neuronal death in several neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis. Over the past few years, we elucidated the mechanisms of D-serine synthesis by the enzyme serine racemase and discovered novel pathways regulating D-serine production and release from neural cells. We also discovered that D-serine is the dominant NMDAR co-agonist mediating neurotoxicity, raising the possibility that drugs that curb D-serine synthesis or release might be useful in neurodegenerative diseases involving NMDARs over-stimulation. We use molecular biology, biochemical and cell biology techniques in tissues and cell culture models. We generate and keep new mouse genetic models of brain transporters and metabolic enzymes, and also apply neurobiological techniques both in vitro and in vivo.
Neuroscience
The Neuroethological Laboratory of Associate Professor Yoram Gutfreund.
Our research questions relate to the evolution and brain mechanisms of cognitive functions such as memory, sensory perception, attention and navigation. Our approach is twofold: Comparative – studying two bird species, the barn owl and the domestic quail, and Neuroethological – progressing from behavior to the underlying neural mechanisms. We use cutting-edge technology to record from large populations of neurons in freely behaving and flying birds. We aim to understand the interface between the physiology of the brain and animal behaviour. Our group is composed of enthusiastic students with various backgrounds (engineering, biology, physics etc.).
__________________________________________________________________________________________________
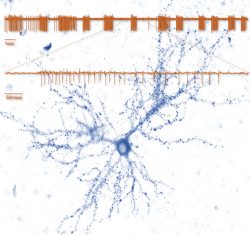 The Synaptic Tenacity and Maintenance Laboratory of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
The Synaptic Tenacity and Maintenance Laboratory of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
__________________________________________________________________________________________________________________
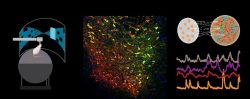 The Neural circuits for Complex Behavior Laboratory of Assistant Professor Ben Engelhard. How do neural circuits in the brain allow us to make decisions, interact with each other, and learn? We are a new lab that investigates the neural activity underlying complex behavior in mice and focus on the brain’s dopamine system, which is essential to many cognitive processes such as decision-making, reward-based learning and social behavior. We use state-of-the-art tools including multiphoton imaging, a virtual-reality behavioral system for mice, optogenetic brain manipulations, and mathematical modeling. We are currently seeking highly motivated graduate students to join the lab. Projects in the lab are varied and suitable to a wide variety of backgrounds (engineering, biology, physics, and more), but enthusiasm for brain research is a must. To inquire please email benengelhard@technion.ac.il with your CV and a short statement of research interests.
The Neural circuits for Complex Behavior Laboratory of Assistant Professor Ben Engelhard. How do neural circuits in the brain allow us to make decisions, interact with each other, and learn? We are a new lab that investigates the neural activity underlying complex behavior in mice and focus on the brain’s dopamine system, which is essential to many cognitive processes such as decision-making, reward-based learning and social behavior. We use state-of-the-art tools including multiphoton imaging, a virtual-reality behavioral system for mice, optogenetic brain manipulations, and mathematical modeling. We are currently seeking highly motivated graduate students to join the lab. Projects in the lab are varied and suitable to a wide variety of backgrounds (engineering, biology, physics, and more), but enthusiasm for brain research is a must. To inquire please email benengelhard@technion.ac.il with your CV and a short statement of research interests.
_____________________________________________________________________________________________________________________
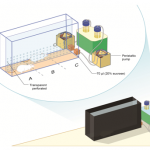 The Behavioral Neuroscience Laboratory of Assistant Professor Avraham (Avi) Avital. In my lab we study behavioral and physiological aspects of attention and the ability to socially cooperate along the translational axis. Acknowledging the crucial role of attention in social as well as in many cognitive processes in health and disease, we are investigating sustained attention using animal-technology interfaces to better understand its neurophysiological mechanisms. Concomitantly, this basic research in rat models is translated to studies in working-dogs, and ends in human research. Specifically, we have developed a software that interfaces with EMG instrument to examine an auditory sustained attention in normal as well as abnormal conditions (e.g; ADHD, PTSD, fatigue, etc.). The importance of social cooperation research stems from a variety of neuropsychiatric disorders that characterized by disruptions in social behavior including: psychopathic personality disorder and Alzheimer’s disease. Developing a fully automated maze, we are able to investigate the gene-environment mechanism of social cooperation.
The Behavioral Neuroscience Laboratory of Assistant Professor Avraham (Avi) Avital. In my lab we study behavioral and physiological aspects of attention and the ability to socially cooperate along the translational axis. Acknowledging the crucial role of attention in social as well as in many cognitive processes in health and disease, we are investigating sustained attention using animal-technology interfaces to better understand its neurophysiological mechanisms. Concomitantly, this basic research in rat models is translated to studies in working-dogs, and ends in human research. Specifically, we have developed a software that interfaces with EMG instrument to examine an auditory sustained attention in normal as well as abnormal conditions (e.g; ADHD, PTSD, fatigue, etc.). The importance of social cooperation research stems from a variety of neuropsychiatric disorders that characterized by disruptions in social behavior including: psychopathic personality disorder and Alzheimer’s disease. Developing a fully automated maze, we are able to investigate the gene-environment mechanism of social cooperation.
 The Theoretical Neuroscience Laboratory of Assistant Professor Omri Barak. What is the nature of neural representation? How does the activity of neurons in the brain reflect the external stimuli and internal states of the animal? Even the simplest of behaviors utilizes large populations of neurons. Thus it seems unlikely that single neurons will exhibit clear and simple relationships to external stimuli or internal states. Our ability to record the detailed activity of single neurons, however, has led to a bias in searching for precisely these types of neurons. Recent advances in recording from larger populations of neurons in an unbiased manner reveal that complex response properties seem to be the rule, and simple neurons the exception. Using theoretical tools from the fields of Mathematics and Physics, along with analysis of experimental data from collaborating laboratories, my lab aims to understand the nature of neural representations.
The Theoretical Neuroscience Laboratory of Assistant Professor Omri Barak. What is the nature of neural representation? How does the activity of neurons in the brain reflect the external stimuli and internal states of the animal? Even the simplest of behaviors utilizes large populations of neurons. Thus it seems unlikely that single neurons will exhibit clear and simple relationships to external stimuli or internal states. Our ability to record the detailed activity of single neurons, however, has led to a bias in searching for precisely these types of neurons. Recent advances in recording from larger populations of neurons in an unbiased manner reveal that complex response properties seem to be the rule, and simple neurons the exception. Using theoretical tools from the fields of Mathematics and Physics, along with analysis of experimental data from collaborating laboratories, my lab aims to understand the nature of neural representations.
The Synaptic Plasticity Laboratory of Senior Lecturer Shai Berlin. Study of synaptic plasticity using novel Optogenetics tools. We are interested in the molecular mechanisms governing synaptic plasticity and memory formation. How is information stored in neurons, in particular in dendritic spines? Spines are small, mushroom-like protrusions on dendrites that encompass the excitatory synapse, as well as a hundred more molecules and proteins. Biochemical and structural changes in the spine are thought to mediate ‘information’ storage. What are the changes that enable a spine to remember? How long can these changes last? Can these be modulated/controlled? To answer these questions, we employ a variety ofmethods ranging from molecular biology through electrophysiology and advanced imaging methods to optogenetics, to name a few. In particular, we design our own tools (optical and/or genetic) to monitor and control cellular processes to better understand synaptic plasticity.
 The Spatial Perception and Memory Laboratory of Associate Professor Dori Derdikman. How is space represented in the brain? How do we form our spatial memory? How are space and time related in the brain? What is the neuronal code for space? The Derdikman lab aims at researching these and related questions. We use diverse methods to conduct neuroscience research, including state-of-the-art electrophysiological recordings using tetrodes from awake-behaving animals, both tethered and telemetry-based, virtual-reality setups for animals, optogenetic methods, and mathematical modeling. The multi-disciplinary work in our lab hosts collaborations between biologists, electrical engineers, medical researchers, physicists, computer scientists, and mathematicians, all under the umbrella of brain science. We record neurons in the brain’s spatial cognitive map system, including grid cells in the entorhinal cortex, place cells in the hippocampus, and head-direction cells. We strive to understand the neural networks that encode our perception and dictate the animal’s behavior in such spaces.
The Spatial Perception and Memory Laboratory of Associate Professor Dori Derdikman. How is space represented in the brain? How do we form our spatial memory? How are space and time related in the brain? What is the neuronal code for space? The Derdikman lab aims at researching these and related questions. We use diverse methods to conduct neuroscience research, including state-of-the-art electrophysiological recordings using tetrodes from awake-behaving animals, both tethered and telemetry-based, virtual-reality setups for animals, optogenetic methods, and mathematical modeling. The multi-disciplinary work in our lab hosts collaborations between biologists, electrical engineers, medical researchers, physicists, computer scientists, and mathematicians, all under the umbrella of brain science. We record neurons in the brain’s spatial cognitive map system, including grid cells in the entorhinal cortex, place cells in the hippocampus, and head-direction cells. We strive to understand the neural networks that encode our perception and dictate the animal’s behavior in such spaces.
The Brain Systems Organization in Health and Disease Laboratory of Associate Professor Itamar Kahn. In my lab we seek to understand how systems-level dynamics give rise to various aspects of brain function and behavior in health and disease. To advance these goals, we take advantage of whole-brain functional imaging in humans and animal models. Functional magnetic resonance imaging (fMRI) allows us to measure activity in multiple brain systems simultaneously, and to look at dynamic interactions between regions of the brain. To manipulate well defined populations of neurons, we are using optogenetic techniques that enable cell-type specific optical control of electrical activity at a millisecond resolution. Combining optogenetic techniques with whole-brain fMRI (termed opto-fMRI), we study the mechanisms governing neural dynamics at the level of the microcircuit and across brain
regions.
 The Cortical Computation Lab of Professor Jackie Schiller. In my lab we investigate the fundamental mechanisms of information processing in dendrites of cortical neurons, and their possible contribution to the network function. In addition, we study the cellular mechanisms of memory and learning. Sensitive optical and electrophysiological tools are used to visualize and monitor neuronal function in the intact tissue. The lab is currently seeking a highly motivated graduate student or post-doc to lead projects dealing with questions related to the cellular basis of sensory processing and plasticity using combined microscopy, electrophysiology, histology, optogenetics and modeling methods both in-vitro in the slice preparation and in-vivo combining behavioral paradigms. For more details call 04-8295270 or send an e-mail to jackie@technion.ac.il.
The Cortical Computation Lab of Professor Jackie Schiller. In my lab we investigate the fundamental mechanisms of information processing in dendrites of cortical neurons, and their possible contribution to the network function. In addition, we study the cellular mechanisms of memory and learning. Sensitive optical and electrophysiological tools are used to visualize and monitor neuronal function in the intact tissue. The lab is currently seeking a highly motivated graduate student or post-doc to lead projects dealing with questions related to the cellular basis of sensory processing and plasticity using combined microscopy, electrophysiology, histology, optogenetics and modeling methods both in-vitro in the slice preparation and in-vivo combining behavioral paradigms. For more details call 04-8295270 or send an e-mail to jackie@technion.ac.il.
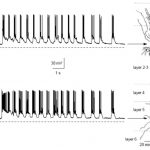 The Epliepsy Research Lab of Associate Professor Yitzhak Schiller. The laboratory’s main research interests are neural network computations, information processing in the motor cortex and basal ganglia, mechanisms underlying ictogenesis in epilepsy, and development of novel neurostimulators and chemo-genetic based treatments for intractable epilepsy. The lab is seeking highly motivated graduate students to lead projects aimed at understanding the pathophysiology of the motor system (in particular in Parkinson’s disease) and epilepsy as well as test novel therapeutic approaches. For more details call 04-8295270 or send an e-mail to syitzhak@technion.ac.il.
The Epliepsy Research Lab of Associate Professor Yitzhak Schiller. The laboratory’s main research interests are neural network computations, information processing in the motor cortex and basal ganglia, mechanisms underlying ictogenesis in epilepsy, and development of novel neurostimulators and chemo-genetic based treatments for intractable epilepsy. The lab is seeking highly motivated graduate students to lead projects aimed at understanding the pathophysiology of the motor system (in particular in Parkinson’s disease) and epilepsy as well as test novel therapeutic approaches. For more details call 04-8295270 or send an e-mail to syitzhak@technion.ac.il.
Physiology, Biophysics, and Systems Biology
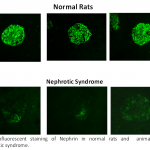 The Vascular Biology Laboratory of Associate Professor Zaid Abassi. Mechanisms of sodium/water retention and cardiac hypertrophy in congestive heart failure. Involvement of the endothelin and nitric oxide systems in the pathogenesis of cardiovascular and metabolic diseases. Pathogenesis of proteinuria in experimental models of nephrotic syndrome: Novel therapeutic approaches. Adverse renal effects of pneumoperitoneum: Mechanisms and therapeutic approaches. Acute kidney injury-Pathogenesis, detection and therapy.
The Vascular Biology Laboratory of Associate Professor Zaid Abassi. Mechanisms of sodium/water retention and cardiac hypertrophy in congestive heart failure. Involvement of the endothelin and nitric oxide systems in the pathogenesis of cardiovascular and metabolic diseases. Pathogenesis of proteinuria in experimental models of nephrotic syndrome: Novel therapeutic approaches. Adverse renal effects of pneumoperitoneum: Mechanisms and therapeutic approaches. Acute kidney injury-Pathogenesis, detection and therapy.
The Genome Structure and Function Laboratoryof Assistant Professor Noam Kaplan studies the profound connection between genetic information and its physical organization. We use a combination of advanced computational and experimental methods to decipher how the genome encodes its 3D organization and how this mediates biological function across different biological systems and in disease.
Biophysics & Systems Biology Laboratory of Assistant Professor Yonatan Savir focuses on machineries by which cells sense their environment and accordingly regulate their growth and metabolism; the impacts of age on signal processing networks in cells; information processing in biological systems.
The laboratory of Professor Shimon Marom
Concrete Research Interest: Biophysical and functional aspects of bio-electricity in proteins, cells and networks, focusing on mechanisms underlying emergence, dynamics and adaptation of bioelectrical phenomena at extended timescales.
Broader Interest: Advancing a framework of relational physiology, inspired by old and more recent ideas of relationists in a range of fields, from the science of self-organization and adaptivity, through theoretical biology, to psychology and anthropology. Within the physiological context, a relational approach entails focusing on the functional organization of systems that are embedded in a responsive and adaptive environment, analyzing processes rather than objects, implementing natural input statistics and closed-loop experimental designs.
Immunity, Microbiology and Inflammation
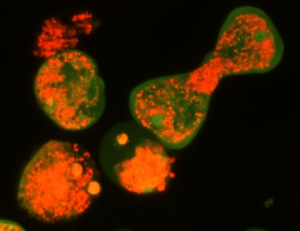 The Emerging Parasites Research Laboratory of Associate Professor Serge Ankri. Infections by parasitic protozoans are largely neglected. With descriptions of the human microbiota, new frontiers of investigation are developing to decipher the complexity of host–parasite–bacteria relationships. Amebiasis, a worldwide intestinal parasitic disease, is due to Entamoeba histolytica. In endemic areas, mixed intestinal infections of E. histolyticaand enteropathogenic Escherichia coliare common. Our laboratory has shown that Escherichia coliO55, protects the parasite against oxidative stress (OS) and induces transcriptome and redox-proteome changes in E.histolytica. Aiming to provide new strategies against amebiasis, we are studying the contribution to OS resistance and proficient intestinal colonization ofE. histolyticaand bacterial factors identified by omics.
The Emerging Parasites Research Laboratory of Associate Professor Serge Ankri. Infections by parasitic protozoans are largely neglected. With descriptions of the human microbiota, new frontiers of investigation are developing to decipher the complexity of host–parasite–bacteria relationships. Amebiasis, a worldwide intestinal parasitic disease, is due to Entamoeba histolytica. In endemic areas, mixed intestinal infections of E. histolyticaand enteropathogenic Escherichia coliare common. Our laboratory has shown that Escherichia coliO55, protects the parasite against oxidative stress (OS) and induces transcriptome and redox-proteome changes in E.histolytica. Aiming to provide new strategies against amebiasis, we are studying the contribution to OS resistance and proficient intestinal colonization ofE. histolyticaand bacterial factors identified by omics.
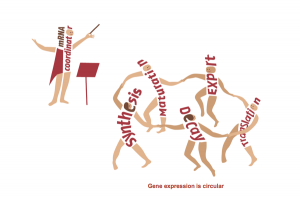 The Regulation of Gene Expression Lab of Professor Mordechai (Motti) Choder.Regulation of gene expression is the process that controls life and determines our phenotype in health and disease. Understanding this process is therefore the focus of research of thousands of groups across the world. Gene expression is traditionally viewed as a linear process divided into distinct stages (e.g., transcription, translation). We have shown that this view is oversimplified. Using the state of the art technologies, we found that all these stages communicate with each other. We are working on factors that shuttle between all these stages and integrate all of them into a system. Among other activities these factors also stimulate mRNA synthesis and degradation. Hence we proposed that gene expression is a circular process – a concept illustrated in the figure. Our work initiated a new direction in studying gene expression that highlight novel targets for controlling cell phenotype and functionality. See http://choder.net.technion.ac.il/research for further information.
The Regulation of Gene Expression Lab of Professor Mordechai (Motti) Choder.Regulation of gene expression is the process that controls life and determines our phenotype in health and disease. Understanding this process is therefore the focus of research of thousands of groups across the world. Gene expression is traditionally viewed as a linear process divided into distinct stages (e.g., transcription, translation). We have shown that this view is oversimplified. Using the state of the art technologies, we found that all these stages communicate with each other. We are working on factors that shuttle between all these stages and integrate all of them into a system. Among other activities these factors also stimulate mRNA synthesis and degradation. Hence we proposed that gene expression is a circular process – a concept illustrated in the figure. Our work initiated a new direction in studying gene expression that highlight novel targets for controlling cell phenotype and functionality. See http://choder.net.technion.ac.il/research for further information.
 The Molecular Virology Laboratory of Associate Professor Tamar Kleinberger. Research in the lab focuses on virus-host cell interactions and on the use of viral proteins for induction of cancer-specific cell death. Specifically, the lab currently studies the mechanisms involved in the ability of the adenovirus E4orf4 protein to inhibit the DNA damage response, a cellular network that identifies DNA damage and transmits signals that promote its repair. This system serves as an antiviral defense mechanism and must be neutralized to improve virus replication. The lab also investigates how E4orf4 and its cellular partners (in particular protein phosphatase 2A and chromatin factors) induce cell death. Understanding these mechanisms may provide an explanation for the cancer specificity of E4orf4-induced cell death and may, in the long run, suggest novel targets for cancer therapy.
The Molecular Virology Laboratory of Associate Professor Tamar Kleinberger. Research in the lab focuses on virus-host cell interactions and on the use of viral proteins for induction of cancer-specific cell death. Specifically, the lab currently studies the mechanisms involved in the ability of the adenovirus E4orf4 protein to inhibit the DNA damage response, a cellular network that identifies DNA damage and transmits signals that promote its repair. This system serves as an antiviral defense mechanism and must be neutralized to improve virus replication. The lab also investigates how E4orf4 and its cellular partners (in particular protein phosphatase 2A and chromatin factors) induce cell death. Understanding these mechanisms may provide an explanation for the cancer specificity of E4orf4-induced cell death and may, in the long run, suggest novel targets for cancer therapy.
 The Yeast Genetics Laboratory of Associate Professor Daniel Kornitzer. We investigate the pathogenic yeast Candida albicans, a prevalent cause of life-threatening infections in immunocompromised patients. We study the adaptations that turned this organism into a successful invader of patients’ blood and tissues, and focus, at the molecular level, on two of its pathogenic characteristics in particular. One is the ability of this fungus to utilize hemoglobin as an iron source, by relying on a newly identified family of extracellular heme-binding proteins. The second is its ability to switch from the yeast morphology to the more invasive mold morphology, and how this switch can be inhibited, i.a.via the activity of the ubiquitin system.
The Yeast Genetics Laboratory of Associate Professor Daniel Kornitzer. We investigate the pathogenic yeast Candida albicans, a prevalent cause of life-threatening infections in immunocompromised patients. We study the adaptations that turned this organism into a successful invader of patients’ blood and tissues, and focus, at the molecular level, on two of its pathogenic characteristics in particular. One is the ability of this fungus to utilize hemoglobin as an iron source, by relying on a newly identified family of extracellular heme-binding proteins. The second is its ability to switch from the yeast morphology to the more invasive mold morphology, and how this switch can be inhibited, i.a.via the activity of the ubiquitin system.
 The Developmental Immunology and Aging Laboratory of Professor Doron Melamed. which focuses on studying mechanisms controlling generation of normal B-lymphocytes and B cell malignancies, to understand how cellular pathways that include signaling molecules, transcription factors and microRNAs operate during B cell development and control crucial fate decisions of normal and transformed B cells. The lab is specifically interested in finding how sensitivity and resistance to cell death signals are regulated in these cells. An additional topic of interest is in investigating how homeostatic mechanisms control B lymphopiesis in general, and particularly in aging, though attempts to identify cross-talk mediators between peripheral B cells and progenitors in the bone marrow that can be targeted to restore immune competence in aging.
The Developmental Immunology and Aging Laboratory of Professor Doron Melamed. which focuses on studying mechanisms controlling generation of normal B-lymphocytes and B cell malignancies, to understand how cellular pathways that include signaling molecules, transcription factors and microRNAs operate during B cell development and control crucial fate decisions of normal and transformed B cells. The lab is specifically interested in finding how sensitivity and resistance to cell death signals are regulated in these cells. An additional topic of interest is in investigating how homeostatic mechanisms control B lymphopiesis in general, and particularly in aging, though attempts to identify cross-talk mediators between peripheral B cells and progenitors in the bone marrow that can be targeted to restore immune competence in aging.
 The Sleep and Psychoneuroimmunology Laboratory of Associate Professor Asya Rolls. Research in the lab focuses on the interaction between the emotional state and physical well-being. Using state of the art tools in neuroscience and in immunology the lab studies the neuronal basis of brain-immune communication, and in particular on two major aspects of brain activity; first, how does sleep and sleep disorders affect the immune system and second, how neuronal networks involved in positive emotions alter the ability of the immune system to fight infections and cancer.
The Sleep and Psychoneuroimmunology Laboratory of Associate Professor Asya Rolls. Research in the lab focuses on the interaction between the emotional state and physical well-being. Using state of the art tools in neuroscience and in immunology the lab studies the neuronal basis of brain-immune communication, and in particular on two major aspects of brain activity; first, how does sleep and sleep disorders affect the immune system and second, how neuronal networks involved in positive emotions alter the ability of the immune system to fight infections and cancer.
 Systems Immunology and Precision Medicine Lab of Assistant Professor Shai Shen-Orr. The immune system is at the heart of the majority of diseases. Yet, albeit extreme disease states, our current understanding of how the immune system varies over time and between individuals is close to nil. We hypothesize that the variability we observe among individuals forms a complex continuum of system states whose constraints we still do not understand, nor how it is affected by external (environmental) or internal (genetics) variability. To chart the immune system landscape and understand the forces shaping it, we are undertaking holistic studies, in which we comprehensively measure the immune system, predominantly its adaptation to chronic conditions. Specifically, in immune aging, inflammatory bowel disease and the inter-cellular interaction network governing cellular communication, including with the brain. These studies both inform our basic understanding of how emergent proprties of immunity arise as well as deliver actionable information to better precision medicine. Students in the lab come from diverse backgrounds, including biology, biotechnology, computer science and physics.
Systems Immunology and Precision Medicine Lab of Assistant Professor Shai Shen-Orr. The immune system is at the heart of the majority of diseases. Yet, albeit extreme disease states, our current understanding of how the immune system varies over time and between individuals is close to nil. We hypothesize that the variability we observe among individuals forms a complex continuum of system states whose constraints we still do not understand, nor how it is affected by external (environmental) or internal (genetics) variability. To chart the immune system landscape and understand the forces shaping it, we are undertaking holistic studies, in which we comprehensively measure the immune system, predominantly its adaptation to chronic conditions. Specifically, in immune aging, inflammatory bowel disease and the inter-cellular interaction network governing cellular communication, including with the brain. These studies both inform our basic understanding of how emergent proprties of immunity arise as well as deliver actionable information to better precision medicine. Students in the lab come from diverse backgrounds, including biology, biotechnology, computer science and physics.
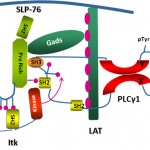 The Immune Cell Signaling, Migration and Cancer Laboratory of Assistant Professor Debbie Yablonski. Immune cells are remarkable for their extraordinary sensitivity to antigen, coupled with extreme selectivity. The lab studies the molecular regulatory mechanisms that underly this behavior, to explain how immune cells are able to mount potent responses to minute quantities of foreign antigens, while ignoring related “self” antigens. This question is studied in T cells, the central arm of the cellular immune system, and in mast cells, an important mediator of allergy. Within the cells, tightly regulated signal processing mechanisms are employed to integrate and interpret information about the presence of antigens, resulting in a cellular decision about how to respond. Specialized signaling proteins, known as adaptors, play an essential role in this process, and are the focus of the lab’s research. A variety of genetic approaches, including mouse models and somatic cell genome editing techniques, are used by the lab to explore the ways in which adaptor proteins fine tune immune sensitivity and selectivity.
The Immune Cell Signaling, Migration and Cancer Laboratory of Assistant Professor Debbie Yablonski. Immune cells are remarkable for their extraordinary sensitivity to antigen, coupled with extreme selectivity. The lab studies the molecular regulatory mechanisms that underly this behavior, to explain how immune cells are able to mount potent responses to minute quantities of foreign antigens, while ignoring related “self” antigens. This question is studied in T cells, the central arm of the cellular immune system, and in mast cells, an important mediator of allergy. Within the cells, tightly regulated signal processing mechanisms are employed to integrate and interpret information about the presence of antigens, resulting in a cellular decision about how to respond. Specialized signaling proteins, known as adaptors, play an essential role in this process, and are the focus of the lab’s research. A variety of genetic approaches, including mouse models and somatic cell genome editing techniques, are used by the lab to explore the ways in which adaptor proteins fine tune immune sensitivity and selectivity.
Clinical Research
The Psychobiology Research Laboratory of Lecturer Alon Shamir, Mazor Mental health center – Acre. The laboratory of Dr. Shamir focuses on elucidating the behavioral, biochemical and molecular consequences of the NRG/ErbB signaling pathway in complex behaviors related to schizophrenia and other psychiatric disorder, on underlying the therapeutics potential of the pathway, and on the identification of biomarkers for schizophrenia.
Infectious Diseases Institute at the Rambam Health Care Campus of Associate Professor Michal Paul. Clinical / epidemiological studies in infectious diseases, multidrug-resistant bacteria, antibiotic therapy, systematic review/ meta-analyses in infectious diseases.
The Laboratory of Stem Cells Utilization in Diabetic Wound Healing of Assistant Professor Mogher Khamaisi, Rambam Health Care Campus. Research in our laboratory focuses on molecular and cellular mechanisms underlying impaired wound healing in diabetic patients, as well as evaluating the efficacy of patient derived induced pluripotent stem cells (iPSCs) in enhancing diabetic wound healing.
Laboratory for Bone Repair, CRIR of Senior Lecturer Hadar Zigdon Giladi, Rambam Health Care Campus. Research focus – study methods to induce bone regeneration using scaffolds, cells and growth factors. Specifically, we study the ability of endothelial progenitor cells (EPCs) to enhance bone regeneration.
Gynecology Research Laboratory of Associate Professor Moti HalakatHillel Yaffe Medical center.
The laboratory of Clinical Neurophysiology of Prof. David Yarnitsky explores pain processing in humans, in health and in disease. We employ tools such as psychophysical testing, electroencephalography (EEG) and event related potentials (ERP) as well as MRI imaging and psychological profiling in order to understand how pain processing done in health, how is it changed in disease, and how can this understanding help in tailoring individual pain treatment.
Before applying, the applicant must first identify a faculty member that will agree to serve as a thesis advisor. Find below a list of faculty members who are currently seeking M.Sc. students. To apply, please contact Ms. Shiri Shporen at the faculty of medicine graduate admissions office (04-829-5374).
Tuition fees for a magister’s degree are determined based on the study requirements given to the student at the time of admission. The thesis equals 20 credits, a final project equals 12 credits, and course load will be determined based on the academic background of the student. The tuition is determined for the entire degree, where 100% tuition means one year’s tuition. Thus, the tuition for a degree is usually 200-300%. If the student is not seeking a degree, the tuition will be 50% per semester. Upon admission, two forms (Bank form & Declaration form) need to be filled out and submitted to the Tuition Fees Departments.
Students who receive scholarships (most students at the faculty of medicine) are exempt from tuition and receive a monthly stipend (see FINANCIAL SUPPORT below for eligibility).
For details please either e-mail Ms. Shiri Shporen or call at 04-829-5374.
For more information, see the Technion Graduate School website.
Only applicants holding M.Sc. or M.D. degrees and who excelled in their studies (percentile grade point average of 88 or above) will be accepted. The acceptance of applicants with international degrees will be pending approval of the Graduate School of Technion. The decision of the departmental Graduate Studies Committee will be based on grades, curriculum vitae, professional achievements and letters of recommendation. The applicant is expected to find a prospective advisor from the Faculty staff, prior to registration. Forms of “Agreement to Serve as Advisor” are available at the departmental Office for Graduate Studies
Before applying, the applicant must first identify a faculty member that will agree to serve as a thesis advisor. Find below a list of faculty members who are currently seeking Ph.D. students. To apply, please contact Ms. Shiri Shporen at the faculty of medicine graduate admissions office (04-829-5374).
For more information, see the Graduate School website.
Find an Advisor
The applicant is expected to find a prospective advisor prior to registration. Prospective students are encouraged to identify potential faculty members and contact them directly.
If you would like to receive general guidance, you can contact Prof. Oded Lewinson , the graduate program coordinator. For application process questions and an Agreement to Serve as Advisor form, contact Ms. Shiri Shporen, the graduate program administrator.
 My research focuses mainly on inherited retinal degenerations (IRD), a heterogeneous group of diseases, which cause visual loss due to the premature death of photoreceptors in the retina. My research combines genetic and functional studies. I recruit IRD patients and their families, and perform various genetic analyses to identify the genetic cause for disease in each family. Identified mutations are studied at the population level, to identify common founder mutations in certain Israeli ethnic groups. Identified genes are further studied to characterize their retinal expression pattern and function.
My research focuses mainly on inherited retinal degenerations (IRD), a heterogeneous group of diseases, which cause visual loss due to the premature death of photoreceptors in the retina. My research combines genetic and functional studies. I recruit IRD patients and their families, and perform various genetic analyses to identify the genetic cause for disease in each family. Identified mutations are studied at the population level, to identify common founder mutations in certain Israeli ethnic groups. Identified genes are further studied to characterize their retinal expression pattern and function. The Laboratory of Tissue Development of Associate Professor Peleg Hasson. The extracellular matrix and the connective tissues that secrete it play cardinal instructive roles, regulating numerous processes in development and regeneration, health and disease. Although many of the matrix’s components have been identified, understanding of how its building blocks are laid and organized and how these properties affect the underlying cells and tissues are still largely unclear. Using mouse genetics and cell-culture based assays, our lab focuses on two major processes which the matrix and connective tissues contribute to: the development and regeneration of the muscles and tendons as well as the development, maturation and remodeling of the blood vasculature.
The Laboratory of Tissue Development of Associate Professor Peleg Hasson. The extracellular matrix and the connective tissues that secrete it play cardinal instructive roles, regulating numerous processes in development and regeneration, health and disease. Although many of the matrix’s components have been identified, understanding of how its building blocks are laid and organized and how these properties affect the underlying cells and tissues are still largely unclear. Using mouse genetics and cell-culture based assays, our lab focuses on two major processes which the matrix and connective tissues contribute to: the development and regeneration of the muscles and tendons as well as the development, maturation and remodeling of the blood vasculature.
The Evolutionary Process of Mutation & Natural Selection Research Laboratory of Associate Professor Ruth Hershberg. Many medically relevant phenomena are in fact evolutionary processes. These include cancer initiation and progression, pathogen emergence and adaptation and the emergence and spread of antibiotic resistance within bacterial populations. In our lab we aim to use evolutionary insights, tools and perspectives, together with cutting edge molecular, genomic and bioinformatics techniques to study such medically relevant evolutionary processes. More specific areas of research include (but are not limited to): (1) What are the antibiotic-independent fitness effects of antibiotic resistance mutations and how do these affect the spread of resistance within natural bacterial populations? (2) Using evolutionary insights and tools to identify cancer driving genes (3) Pathogen evolution via gene loss.
 The Developmental Genetics Research Laboratory of Associate Professor Adi Salzberg. One of the fascinating questions in developmental biology is how different cells, which develop within a given tissue, acquire different properties that allow them to work together as a functional unit. We are using the proprioceptive organs in the peripheral nervous system (PNS) of Drosophila(also called chordotonal organs, ChOs) as a model system for studying cell fate diversification. The fly ChOs, sensory organs that detect motion and position, are composed of linear arrays of lineage-related cells with distinct identities and properties that allow them to work together as a functional organ. Very little is known about the mechanisms regulating the specification of the various ChO cell identities. Through our work on the deigene we have identified distinct regulatory elements that direct gene expression to specific ChO cell types. We have used these elements to construct a unique transgenic fly strain in which different ChO cell types are labeled with different fluorescent markers. This strain is currently being used to conduct a large-scale, RNAi-based, genetic screen for novel determinants of cell fate specification and morphogenesis in the ChO lineage.
The Developmental Genetics Research Laboratory of Associate Professor Adi Salzberg. One of the fascinating questions in developmental biology is how different cells, which develop within a given tissue, acquire different properties that allow them to work together as a functional unit. We are using the proprioceptive organs in the peripheral nervous system (PNS) of Drosophila(also called chordotonal organs, ChOs) as a model system for studying cell fate diversification. The fly ChOs, sensory organs that detect motion and position, are composed of linear arrays of lineage-related cells with distinct identities and properties that allow them to work together as a functional organ. Very little is known about the mechanisms regulating the specification of the various ChO cell identities. Through our work on the deigene we have identified distinct regulatory elements that direct gene expression to specific ChO cell types. We have used these elements to construct a unique transgenic fly strain in which different ChO cell types are labeled with different fluorescent markers. This strain is currently being used to conduct a large-scale, RNAi-based, genetic screen for novel determinants of cell fate specification and morphogenesis in the ChO lineage.
The Synaptic Tenacity and Maintenance Laboratory of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
___________________________________________________________________________________________________________________
 The Epithelial Stem Cells and Pathophysiology Research Laboratory of Assistant Professor Ruby Shalom-Feuerstein. Our lab explores the molecular events that underlie the morphogenesis, stem cell homeostasis, and pathophysiology of the skin and the cornea. We employ complementary approaches, some of which were uniquely developed in our lab, to investigate these biological processes and develop novel therapies. Among these technologies are induced pluripotent stem cell and adult stem cell cultures, lineage tracing experiments and genetic mouse models, molecular biology, and genome wide analysis.
The Epithelial Stem Cells and Pathophysiology Research Laboratory of Assistant Professor Ruby Shalom-Feuerstein. Our lab explores the molecular events that underlie the morphogenesis, stem cell homeostasis, and pathophysiology of the skin and the cornea. We employ complementary approaches, some of which were uniquely developed in our lab, to investigate these biological processes and develop novel therapies. Among these technologies are induced pluripotent stem cell and adult stem cell cultures, lineage tracing experiments and genetic mouse models, molecular biology, and genome wide analysis.
The Laboratory of Organogenesis and Developmental Biology of Associate Professor Tom Schultheiss.  The fields of Regenerative Medicine and Tissue Engineering, which aim to generate tissues in the laboratory in order to replace and repair damaged organs, hold great promise for treating many congenital and chronic diseases. In order to generate tissues and organs in the lab, it essential to understand how these tissues are normally built in the developing embryo. The Schultheiss laboratory studies how organs are built in the embryo, from the first steps when embryonic cells adopt tissue-specific identities, through the later stages when the functional organ is assembled from its various cellular components. Our current work focuses on the formation of the kidney, gonads, and hematopoietic system, and we use the avian embryo (chicken and quail) as our main experimental system. We also have a strong interest in understanding basic embryological processes, such as embryonic patterning, cell differentiation, migration, and morphogenesis.
The fields of Regenerative Medicine and Tissue Engineering, which aim to generate tissues in the laboratory in order to replace and repair damaged organs, hold great promise for treating many congenital and chronic diseases. In order to generate tissues and organs in the lab, it essential to understand how these tissues are normally built in the developing embryo. The Schultheiss laboratory studies how organs are built in the embryo, from the first steps when embryonic cells adopt tissue-specific identities, through the later stages when the functional organ is assembled from its various cellular components. Our current work focuses on the formation of the kidney, gonads, and hematopoietic system, and we use the avian embryo (chicken and quail) as our main experimental system. We also have a strong interest in understanding basic embryological processes, such as embryonic patterning, cell differentiation, migration, and morphogenesis.
 Laboratory for Mechanobiology of the Cell of Assistant Professor Haguy Wolfenson. Cells are exposed to a variety of signals that control their fate. Whereas biomedical studies are mostly focused on biochemical signals, the mechanical features of the microenvironment around cells are as much important in regulating cellular functions such as migration, survival, growth, and differentiation. The studies in the lab are focused on the mechanisms by which cells sense and respond to mechanical features of their environment such as rigidity, topography, or ligand density. We use a combination of advanced microscopy techniques with biophysical measurements to decipher the mechanisms by which extracellular mechanical signals are transmitted into biochemical signals inside the cells and how those in turn affect cellular behavior. We also study how these processes are altered in cancer with the premise that proper mechanosensing is malfunctioning in cancer cells.
Laboratory for Mechanobiology of the Cell of Assistant Professor Haguy Wolfenson. Cells are exposed to a variety of signals that control their fate. Whereas biomedical studies are mostly focused on biochemical signals, the mechanical features of the microenvironment around cells are as much important in regulating cellular functions such as migration, survival, growth, and differentiation. The studies in the lab are focused on the mechanisms by which cells sense and respond to mechanical features of their environment such as rigidity, topography, or ligand density. We use a combination of advanced microscopy techniques with biophysical measurements to decipher the mechanisms by which extracellular mechanical signals are transmitted into biochemical signals inside the cells and how those in turn affect cellular behavior. We also study how these processes are altered in cancer with the premise that proper mechanosensing is malfunctioning in cancer cells.
The Laboratory of Gene Regulation and Phenotypic Evolution of Assistant Professor Ella Preger-Ben Noon. Our lab studies the molecular mechanisms underlying enhancer function and  evolution. We investigate how mutations in these regulatory DNA sequences lead to the evolution of animal body form. To address these questions, we combine the powerful genetic toolkit of the fruit fly Drosophila with cutting-edge techniques of cell type-specific genomics, single-molecule live imaging and genome editing. Our approach allows us to untangle the precise molecular mechanisms that underlie phenotypic changes between closely related species and to uncover new models of gene regulation.
evolution. We investigate how mutations in these regulatory DNA sequences lead to the evolution of animal body form. To address these questions, we combine the powerful genetic toolkit of the fruit fly Drosophila with cutting-edge techniques of cell type-specific genomics, single-molecule live imaging and genome editing. Our approach allows us to untangle the precise molecular mechanisms that underlie phenotypic changes between closely related species and to uncover new models of gene regulation.
 The role of ATF3/JDP2 bZIP proteins in heart pathophysiology laboratory of Prof. Ami Aronheim. Hypertrophic growth of the cardiac muscle is an adaptive response that occurs in physiological and pathological conditions. However, whereas physiological hypertrophy is beneficial for heart function, sustained pathological hypertrophy leads to cardiac maladaptive remodeling processes resulting in permanent contractile dysfunction and heart failure. Currently, the incidence of heart failure is on the rise and treatment options are limited. Cardiac remodeling is regulated by multiple G-protein coupled receptors that converge to a limited number of transcription factors. Thus, targeting key transcriptional regulators emerges as a promising strategy for the development of new therapeutic approaches. Over the last decade, we have studied the role of the Activating Transcription Factor 3 (ATF3), a basic leucine zipper protein (bZIP), in promoting maladaptive cardiac remodeling processes in response to pressure overload. ATF3 is most closely homologous to the c-Jun Dimerization Protein 2 (JDP2), a bZIP gene that was initially isolated in our lab. ATF3 and JDP2 are primarily transcription inhibitors. Our results show that hearts derived from mice lacking both ATF3 and JDP2 are enlarged, exhibit an improved contractile function, however, with no apparent increase in either hypertrophic, fibrosis or inflammatory markers. In addition, these mice are protected from pressure overload-induced maladaptive cardiac remodeling. We aim to study the molecular mechanisms leading to protection of the heart from maladaptive cardiac remodeling processes. Ultimately, this study will provide new insights towards the development of novel strategies for the treatment of cardiac dysfunction and heart failure.
The role of ATF3/JDP2 bZIP proteins in heart pathophysiology laboratory of Prof. Ami Aronheim. Hypertrophic growth of the cardiac muscle is an adaptive response that occurs in physiological and pathological conditions. However, whereas physiological hypertrophy is beneficial for heart function, sustained pathological hypertrophy leads to cardiac maladaptive remodeling processes resulting in permanent contractile dysfunction and heart failure. Currently, the incidence of heart failure is on the rise and treatment options are limited. Cardiac remodeling is regulated by multiple G-protein coupled receptors that converge to a limited number of transcription factors. Thus, targeting key transcriptional regulators emerges as a promising strategy for the development of new therapeutic approaches. Over the last decade, we have studied the role of the Activating Transcription Factor 3 (ATF3), a basic leucine zipper protein (bZIP), in promoting maladaptive cardiac remodeling processes in response to pressure overload. ATF3 is most closely homologous to the c-Jun Dimerization Protein 2 (JDP2), a bZIP gene that was initially isolated in our lab. ATF3 and JDP2 are primarily transcription inhibitors. Our results show that hearts derived from mice lacking both ATF3 and JDP2 are enlarged, exhibit an improved contractile function, however, with no apparent increase in either hypertrophic, fibrosis or inflammatory markers. In addition, these mice are protected from pressure overload-induced maladaptive cardiac remodeling. We aim to study the molecular mechanisms leading to protection of the heart from maladaptive cardiac remodeling processes. Ultimately, this study will provide new insights towards the development of novel strategies for the treatment of cardiac dysfunction and heart failure.
___________________________________________________________________________________________________________________
 Aaron Ciechanover, MD, DSc Distinguished Professor Laureate, the 2004 Nobel Prize in Chemistry The ubiquitin proteolytic system, pathogenesis of diseases,
Aaron Ciechanover, MD, DSc Distinguished Professor Laureate, the 2004 Nobel Prize in Chemistry The ubiquitin proteolytic system, pathogenesis of diseases,
and development of therapeutic modalities.
Our group is part of the Rappaport – Technion Integrated Cancer Center – R-TICC and we work in tight collaboration with many of its researchers. Our studies focus on the role of the ubiquitin system (that was discovered in our laboratory and was culminated with the awarding of the 2004 Nobel Prize in Chemistry) in basic mechanisms of diseases, cancer and neurodegeneration in particular. Our research is an elegant example of how curiosity-driven basic research leads – by different groups – to its translation to efficient drugs to combat severe human diseases – including malignancies and neurodegeneration.
One area of studies is the mechanism by of activation of NF-κB, a major transcription factor that is upregulated in many tumors. We identified a surprising side to this known story when we showed that overexpression of its p50 subunit, which is typically generated in stoichiometric amounts to the other partner – p65, displays a strong tumor suppressive effect that we now study.
Another area of studies involves the proteasome, which is the catalytic arm of the ubiquitin system. Here we showed that by simple metabolic “trick” we can lock it in the nucleus, preventing it from supplying the cell with amino acids derived from degradation of intracellular proteins, and thus resulting in cell death. It appears that cancer cells are more sensitive to this lock, as their metabolic needs are much larger than those of healthy cells.
In an independent line of research, we are studying the involvement of the ubiquitin system in neurodegenerative disorders – the hallmark of which is accumulation in the brain of abnormal/mutated and typically aggregated proteins that are not recognized by the ubiquitin system. Out of several diseases, we decided to study Huntington’s disease where several good experimental tools – probes, cells, and animal models – are available. Huntington’s disease (HD) is a progressive incurable neurodegenerative disorder characterized by motor and neuropsychiatric symptoms
Cancer Metabolism Laboratory of Professor Eyal Gottlieb. We utilize metabolomics approaches to identify metabolic alterations and to target metabolic essential pathways of cancer cells.
The Computational Cancer Genomics lab of Dr. Yizhak focuses on computational challenges in cancer research. As opposed to traditional “wet” labs, much of the biological and medical  research today is done computationally in what we call “dry” labs. Such research is enabled by large amount of clinical and molecular data collected around the world by the scientific community.
research today is done computationally in what we call “dry” labs. Such research is enabled by large amount of clinical and molecular data collected around the world by the scientific community.
Our lab addresses emerging challenges in computational cancer genomics that require the development and application of innovative computational methods and tools for cancer data analysis. We work closely with clinicians and experimentalists in Israel and around the world to turn data into knowledge and advance cancer treatment and prevention.
 Genetic Networks, Tissue Homeostasis and Cancer Laboratory of Associate Professor Amir Orian (Oryan). My laboratory focuses on understanding fundamental mechanisms and genes of cancer-related genetic networks. These networks are critical for maintenance of the differentiated identity and deregulation of such networks is associated with cancer. We focus on: 1. “Identity network”;, genes that are required to maintain the identity of differentiated cells and serve as a “barrier to tumorigenesis.” 2. NOA-gene network; Non-Oncogene Addiction genes that a essential for cancer cells to cope with the oncogenic stress, are vital for maintaining the tumorous phenotype, but are less critical to un-transformed cells. Thus, they are have the potential to be molecular targets for molecular cancer therapy. Our current projects include: 1. Characterizing the role of SUMO-Targeted-Ubiquitin Ligase proteins in Drosophila development and human cancer; 2. Identifying genes involved in innate immune responses to distinct pathogens; and 3. Delineating the genes that regulate adult gut homeostasis at the genetic and genomic level, with a focus on genes that maintain the balance between progenitor and differentiated cells. Towards these aims we employ advanced genetic and genomic tools to study transcriptional networks using Drosophila genetics and genomics, mammalian cells, mouse-derived intestinal organoids and patient-derived specimens.
Genetic Networks, Tissue Homeostasis and Cancer Laboratory of Associate Professor Amir Orian (Oryan). My laboratory focuses on understanding fundamental mechanisms and genes of cancer-related genetic networks. These networks are critical for maintenance of the differentiated identity and deregulation of such networks is associated with cancer. We focus on: 1. “Identity network”;, genes that are required to maintain the identity of differentiated cells and serve as a “barrier to tumorigenesis.” 2. NOA-gene network; Non-Oncogene Addiction genes that a essential for cancer cells to cope with the oncogenic stress, are vital for maintaining the tumorous phenotype, but are less critical to un-transformed cells. Thus, they are have the potential to be molecular targets for molecular cancer therapy. Our current projects include: 1. Characterizing the role of SUMO-Targeted-Ubiquitin Ligase proteins in Drosophila development and human cancer; 2. Identifying genes involved in innate immune responses to distinct pathogens; and 3. Delineating the genes that regulate adult gut homeostasis at the genetic and genomic level, with a focus on genes that maintain the balance between progenitor and differentiated cells. Towards these aims we employ advanced genetic and genomic tools to study transcriptional networks using Drosophila genetics and genomics, mammalian cells, mouse-derived intestinal organoids and patient-derived specimens.
The Tumor Biology Lab of Professor Vlodavskyfocuses on cell interaction with the extracellular matrix (ECM) and the tumor microenvironment. In the center of this research is an enzyme (heparanase) which degrades a major component (heparan sulfate) of the ECM and thereby releases numerous factors that promote tumor growth, metastasis and angiogenesis. Heparanase-inhibiting compounds and neutralizing antibodies are being developed to inhibit cancer progression and inflammation. Among the issues that are being investigated in the Lab are: 1)How dose heparanase activate macrophages and other cells of the immune system? 2) How does heparanase elicit chemoresistance? 3) Can heparanase inhibitors block tumor progression and chemoresistance driven by host heparanase? 4) What is the role of heparanase-2 in normal physiology? and 5) Do heparanase-1 and heparanse-2 play a role in gene transcription?
 The Redox Biology Laboratory of Associate Professor Benhar Moran focuses on studying the mechanisms whereby redox modifications of cysteine residues regulate protein and cell function. Research projects in the lab use cutting-edge proteomic and biochemical tools to explore the roles of protein redox modifications (such as cysteine nitrosylation) in cellular communication, inflammation, and cancer.
The Redox Biology Laboratory of Associate Professor Benhar Moran focuses on studying the mechanisms whereby redox modifications of cysteine residues regulate protein and cell function. Research projects in the lab use cutting-edge proteomic and biochemical tools to explore the roles of protein redox modifications (such as cysteine nitrosylation) in cellular communication, inflammation, and cancer.
 The Molecular Embryology Laboratory of Associate Professor Dale Frank. My research group studies the initial generation of the body axis occurring during embryonic development. Using amphibians as a model vertebrate system, we investigate the cross-talk between transcription factors and signaling pathways to determine how they induce neural and mesodermal cell fates along the body axis during early embryo development. Frogs, like people, are vertebrates, and the regulation of early embryonic development in all vertebrates (including mammals) is carried out by the same gene products regardless, of species. Thus, the frog (Xenopus laevis) is an excellent system to learn how all vertebrates make their body plan. One of the earliest and most dramatic events in early development is the formation of the body axes. As early embryos, we all start out as a round egg cell, which then rapidly divides into groups of cells. These early stem cells are totipotential and can develop into all cell types of the embryo. At a critical stage, cells commit to specific fates. The cells start migrating, and they elongate to make our typical body plan: the head – tail axis (anterior – posterior), the back – stomach axis (dorsal – ventral) and finally the left – right axis. Cells must know what they are doing along theses body axes. Not only must a cell decide its own identity, a decision to be muscle, blood or nerve for instance. Cells must also define their position along the axis. In the nervous system for example, a cell in the anterior-head makes a forebrain-derived neuron or an eye cell, whereas a nerve cell in the posterior-tail region makes a motor neuron. This location-dependence of cell fate is based on the capability of cells to monitor their position with respect to the developing embryonic axes. In my lab, we are working on the genetic and cellular interactions determining how progenitor like stem-like of the early mesoderm and nervous system acquire these different axial cell fates during early development.
The Molecular Embryology Laboratory of Associate Professor Dale Frank. My research group studies the initial generation of the body axis occurring during embryonic development. Using amphibians as a model vertebrate system, we investigate the cross-talk between transcription factors and signaling pathways to determine how they induce neural and mesodermal cell fates along the body axis during early embryo development. Frogs, like people, are vertebrates, and the regulation of early embryonic development in all vertebrates (including mammals) is carried out by the same gene products regardless, of species. Thus, the frog (Xenopus laevis) is an excellent system to learn how all vertebrates make their body plan. One of the earliest and most dramatic events in early development is the formation of the body axes. As early embryos, we all start out as a round egg cell, which then rapidly divides into groups of cells. These early stem cells are totipotential and can develop into all cell types of the embryo. At a critical stage, cells commit to specific fates. The cells start migrating, and they elongate to make our typical body plan: the head – tail axis (anterior – posterior), the back – stomach axis (dorsal – ventral) and finally the left – right axis. Cells must know what they are doing along theses body axes. Not only must a cell decide its own identity, a decision to be muscle, blood or nerve for instance. Cells must also define their position along the axis. In the nervous system for example, a cell in the anterior-head makes a forebrain-derived neuron or an eye cell, whereas a nerve cell in the posterior-tail region makes a motor neuron. This location-dependence of cell fate is based on the capability of cells to monitor their position with respect to the developing embryonic axes. In my lab, we are working on the genetic and cellular interactions determining how progenitor like stem-like of the early mesoderm and nervous system acquire these different axial cell fates during early development.
 The Laboratory of Neurodegeneration of Associate Prof. Simone Engelender. Parkinson’s disease (PD) is a very common neurodegenerative disease and it is caused by the neuronal accumulation of a protein known as a-synuclein. Our goal is to investigate the mechanisms that lead to the accumulation of a-synuclein in the brain of PD patients. We found that monoubiquitination of a-synuclein leads to its degradation by the proteasome. Nevertheless, the degradation of monoubiquitinated a-synuclein is impaired in PD, leading to the accumulation and aggregation of a-synuclein. More recently, we found that additional post-translational modifications, such as SUMOylation, prevents the proteasomal degradation of a-synuclein and leads to even further accumulation and aggregation of a-synuclein into inclusions, which resemble to the inclusions found in the disease. In addition, we identified several proteins of the monoubiquitination and SUMOylation pathways that are dysfunctional in PD brains, further suggesting their role in a-synuclein homeostasis and PD. We are currently utilizing advanced biochemical and cell biology techniques as well as generating novel PD-related transgenic mice to further investigate the mechanisms that regulate a-synuclein degradation, accumulation and aggregation in PD. In an attempt to decrease a-synuclein levels in the disease, we are also generating compounds to specifically target the a-synuclein monoubiquitination and SUMOylation pathways.
The Laboratory of Neurodegeneration of Associate Prof. Simone Engelender. Parkinson’s disease (PD) is a very common neurodegenerative disease and it is caused by the neuronal accumulation of a protein known as a-synuclein. Our goal is to investigate the mechanisms that lead to the accumulation of a-synuclein in the brain of PD patients. We found that monoubiquitination of a-synuclein leads to its degradation by the proteasome. Nevertheless, the degradation of monoubiquitinated a-synuclein is impaired in PD, leading to the accumulation and aggregation of a-synuclein. More recently, we found that additional post-translational modifications, such as SUMOylation, prevents the proteasomal degradation of a-synuclein and leads to even further accumulation and aggregation of a-synuclein into inclusions, which resemble to the inclusions found in the disease. In addition, we identified several proteins of the monoubiquitination and SUMOylation pathways that are dysfunctional in PD brains, further suggesting their role in a-synuclein homeostasis and PD. We are currently utilizing advanced biochemical and cell biology techniques as well as generating novel PD-related transgenic mice to further investigate the mechanisms that regulate a-synuclein degradation, accumulation and aggregation in PD. In an attempt to decrease a-synuclein levels in the disease, we are also generating compounds to specifically target the a-synuclein monoubiquitination and SUMOylation pathways.
The Molecular Machines laboratory  of Associate Professor Oded Lewinson. Our lab is interested in how molecular machines are built and how they work. We are especially interested in molecular machines that are important for cancer multidrug resistance and for bacterial pathogenesis. We harness this knowledge to identify new targets for drug development.
of Associate Professor Oded Lewinson. Our lab is interested in how molecular machines are built and how they work. We are especially interested in molecular machines that are important for cancer multidrug resistance and for bacterial pathogenesis. We harness this knowledge to identify new targets for drug development.
For example, in 2018 our lab received the NATO Prize for Peace and Security following our discovery that zinc can be used as a highly effective inhibitor of an essential virulence factor of Anthrax (shown here on the right).
Structure of the Bacillus Anthracis
virulence determinant MntBC and
its drug binding sites
 Calcium Signaling Laboratory of Senior Lecturer Raz Palty. Calcium ions are central and ubiquitous intracellular signaling entities. Changes in intracellular calcium levels control fundamental cellular events, from signaling the mitotic birth of a new cell to marking its end during apoptosis. To generate calcium signals, cells rely on an elaborate machinery of ion transport proteins that are strategically positioned in the plasma membrane (PM) and intracellular organelle membranes. The function of calcium transport proteins at these sites is critical for numerous physiological processes including synaptic transmission, muscle contraction, and the immune response. Consequently, several disease states arise due to dysfunction of calcium transport mechanisms including neurological disorders, cardiac disease and immunodeficiency. Our main focus in the Palty lab is to elucidate molecular mechanisms that regulate calcium signaling across the PM, endoplasmic reticulum (ER) and mitochondria with the long-term goal of understanding how calcium signals are generated at these sites under normal physiological conditions and in disease states.
Calcium Signaling Laboratory of Senior Lecturer Raz Palty. Calcium ions are central and ubiquitous intracellular signaling entities. Changes in intracellular calcium levels control fundamental cellular events, from signaling the mitotic birth of a new cell to marking its end during apoptosis. To generate calcium signals, cells rely on an elaborate machinery of ion transport proteins that are strategically positioned in the plasma membrane (PM) and intracellular organelle membranes. The function of calcium transport proteins at these sites is critical for numerous physiological processes including synaptic transmission, muscle contraction, and the immune response. Consequently, several disease states arise due to dysfunction of calcium transport mechanisms including neurological disorders, cardiac disease and immunodeficiency. Our main focus in the Palty lab is to elucidate molecular mechanisms that regulate calcium signaling across the PM, endoplasmic reticulum (ER) and mitochondria with the long-term goal of understanding how calcium signals are generated at these sites under normal physiological conditions and in disease states.
 Protein homeostasis regulatory networks – A systems biology approach to protein homeostasis regulation in stress and neurodegenerative diseases.The lab of Associate Professor Reut Shalgi integrates molecular and computational biology towards the goal of understanding gene expression regulation, and protein homeostasis networks, in stressed human cells, and how they are impaired in neurodegenerative diseases. We mainly focus on Huntington’s disease, and ALS, and use cutting-edge technologies, including NGS transcriptomics and translatomics, protein-protein interaction network mapping, high throughput screening, and big data analysis, to understand the mechanisms of these diseases, and reveal how we can help the cells with their battle against them.
Protein homeostasis regulatory networks – A systems biology approach to protein homeostasis regulation in stress and neurodegenerative diseases.The lab of Associate Professor Reut Shalgi integrates molecular and computational biology towards the goal of understanding gene expression regulation, and protein homeostasis networks, in stressed human cells, and how they are impaired in neurodegenerative diseases. We mainly focus on Huntington’s disease, and ALS, and use cutting-edge technologies, including NGS transcriptomics and translatomics, protein-protein interaction network mapping, high throughput screening, and big data analysis, to understand the mechanisms of these diseases, and reveal how we can help the cells with their battle against them.
 The Molecular Neuroscience Laboratory of Professor Herman Wolosker. Research in our laboratory focuses on understanding the roles of unconventional neurotransmitters, like the D-amino acids in the central nervous system. We are particularly interested in the regulation of N-Methyl- D-aspartate receptors (NMDARs) by D-serine, a D-enantiomer previously thought to be restricted to bacteria or lower invertebrates. D-Serine is now increasingly appreciated as a major physiologic ligand for NMDARs that mediates NMDAR synaptic responses and neurotoxicity both in vitro and in vivo. In addition to neurotransmission, NMDARs play a key role in neurodegeneration, with their excessive activation contributing to neuronal death in several neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis. Over the past few years, we elucidated the mechanisms of D-serine synthesis by the enzyme serine racemase and discovered novel pathways regulating D-serine production and release from neural cells. We also discovered that D-serine is the dominant NMDAR co-agonist mediating neurotoxicity, raising the possibility that drugs that curb D-serine synthesis or release might be useful in neurodegenerative diseases involving NMDARs over-stimulation. We use molecular biology, biochemical and cell biology techniques in tissues and cell culture models. We generate and keep new mouse genetic models of brain transporters and metabolic enzymes, and also apply neurobiological techniques both in vitro and in vivo.
The Molecular Neuroscience Laboratory of Professor Herman Wolosker. Research in our laboratory focuses on understanding the roles of unconventional neurotransmitters, like the D-amino acids in the central nervous system. We are particularly interested in the regulation of N-Methyl- D-aspartate receptors (NMDARs) by D-serine, a D-enantiomer previously thought to be restricted to bacteria or lower invertebrates. D-Serine is now increasingly appreciated as a major physiologic ligand for NMDARs that mediates NMDAR synaptic responses and neurotoxicity both in vitro and in vivo. In addition to neurotransmission, NMDARs play a key role in neurodegeneration, with their excessive activation contributing to neuronal death in several neurodegenerative disorders, such as Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis. Over the past few years, we elucidated the mechanisms of D-serine synthesis by the enzyme serine racemase and discovered novel pathways regulating D-serine production and release from neural cells. We also discovered that D-serine is the dominant NMDAR co-agonist mediating neurotoxicity, raising the possibility that drugs that curb D-serine synthesis or release might be useful in neurodegenerative diseases involving NMDARs over-stimulation. We use molecular biology, biochemical and cell biology techniques in tissues and cell culture models. We generate and keep new mouse genetic models of brain transporters and metabolic enzymes, and also apply neurobiological techniques both in vitro and in vivo.
The Neuroethological Laboratory of Associate Professor Yoram Gutfreund.
Our research questions relate to the evolution and brain mechanisms of cognitive functions such as memory, sensory perception, attention and navigation. Our approach is twofold: Comparative – studying two bird species, the barn owl and the domestic quail, and Neuroethological – progressing from behavior to the underlying neural mechanisms. We use cutting-edge technology to record from large populations of neurons in freely behaving and flying birds. We aim to understand the interface between the physiology of the brain and animal behaviour. Our group is composed of enthusiastic students with various backgrounds (engineering, biology, physics etc.).
__________________________________________________________________________________________________
 The Synaptic Tenacity and Maintenance Laboratory of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
The Synaptic Tenacity and Maintenance Laboratory of Professor Noam Ziv. Memories are widely believed to be embedded in neuronal networks though changes in synapses, cell-to-cell contacts specialized for signal transmission. Synapses, however, are tiny, composed of remarkably dynamic constituents which have finite life spans. How do synapses maintain their individual properties over behavioral time scales? How well do they do so? What happens to memories when they don’t? To address these questions, we use advanced imaging techniques, multielectrode array recordings and proteomics to study synaptic tenacity and maintenance, and how these relate to forgetting in physiological settings and in respect to aging and pathological states. Our lab, which is part of the highly multi-disciplinary Network Biology Research center at the Faculty of Electrical & Computer Engineering in the Neve Shaanan campus, is currently looking for a graduate student for a project focusing on synapse maintenance in aging and in the context of neurodegenerative disease.
__________________________________________________________________________________________________________________
 The Neural circuits for Complex Behavior Laboratory of Assistant Professor Ben Engelhard. How do neural circuits in the brain allow us to make decisions, interact with each other, and learn? We are a new lab that investigates the neural activity underlying complex behavior in mice and focus on the brain’s dopamine system, which is essential to many cognitive processes such as decision-making, reward-based learning and social behavior. We use state-of-the-art tools including multiphoton imaging, a virtual-reality behavioral system for mice, optogenetic brain manipulations, and mathematical modeling. We are currently seeking highly motivated graduate students to join the lab. Projects in the lab are varied and suitable to a wide variety of backgrounds (engineering, biology, physics, and more), but enthusiasm for brain research is a must. To inquire please email benengelhard@technion.ac.il with your CV and a short statement of research interests.
The Neural circuits for Complex Behavior Laboratory of Assistant Professor Ben Engelhard. How do neural circuits in the brain allow us to make decisions, interact with each other, and learn? We are a new lab that investigates the neural activity underlying complex behavior in mice and focus on the brain’s dopamine system, which is essential to many cognitive processes such as decision-making, reward-based learning and social behavior. We use state-of-the-art tools including multiphoton imaging, a virtual-reality behavioral system for mice, optogenetic brain manipulations, and mathematical modeling. We are currently seeking highly motivated graduate students to join the lab. Projects in the lab are varied and suitable to a wide variety of backgrounds (engineering, biology, physics, and more), but enthusiasm for brain research is a must. To inquire please email benengelhard@technion.ac.il with your CV and a short statement of research interests.
_____________________________________________________________________________________________________________________
 The Behavioral Neuroscience Laboratory of Assistant Professor Avraham (Avi) Avital. In my lab we study behavioral and physiological aspects of attention and the ability to socially cooperate along the translational axis. Acknowledging the crucial role of attention in social as well as in many cognitive processes in health and disease, we are investigating sustained attention using animal-technology interfaces to better understand its neurophysiological mechanisms. Concomitantly, this basic research in rat models is translated to studies in working-dogs, and ends in human research. Specifically, we have developed a software that interfaces with EMG instrument to examine an auditory sustained attention in normal as well as abnormal conditions (e.g; ADHD, PTSD, fatigue, etc.). The importance of social cooperation research stems from a variety of neuropsychiatric disorders that characterized by disruptions in social behavior including: psychopathic personality disorder and Alzheimer’s disease. Developing a fully automated maze, we are able to investigate the gene-environment mechanism of social cooperation.
The Behavioral Neuroscience Laboratory of Assistant Professor Avraham (Avi) Avital. In my lab we study behavioral and physiological aspects of attention and the ability to socially cooperate along the translational axis. Acknowledging the crucial role of attention in social as well as in many cognitive processes in health and disease, we are investigating sustained attention using animal-technology interfaces to better understand its neurophysiological mechanisms. Concomitantly, this basic research in rat models is translated to studies in working-dogs, and ends in human research. Specifically, we have developed a software that interfaces with EMG instrument to examine an auditory sustained attention in normal as well as abnormal conditions (e.g; ADHD, PTSD, fatigue, etc.). The importance of social cooperation research stems from a variety of neuropsychiatric disorders that characterized by disruptions in social behavior including: psychopathic personality disorder and Alzheimer’s disease. Developing a fully automated maze, we are able to investigate the gene-environment mechanism of social cooperation.
 The Theoretical Neuroscience Laboratory of Assistant Professor Omri Barak. What is the nature of neural representation? How does the activity of neurons in the brain reflect the external stimuli and internal states of the animal? Even the simplest of behaviors utilizes large populations of neurons. Thus it seems unlikely that single neurons will exhibit clear and simple relationships to external stimuli or internal states. Our ability to record the detailed activity of single neurons, however, has led to a bias in searching for precisely these types of neurons. Recent advances in recording from larger populations of neurons in an unbiased manner reveal that complex response properties seem to be the rule, and simple neurons the exception. Using theoretical tools from the fields of Mathematics and Physics, along with analysis of experimental data from collaborating laboratories, my lab aims to understand the nature of neural representations.
The Theoretical Neuroscience Laboratory of Assistant Professor Omri Barak. What is the nature of neural representation? How does the activity of neurons in the brain reflect the external stimuli and internal states of the animal? Even the simplest of behaviors utilizes large populations of neurons. Thus it seems unlikely that single neurons will exhibit clear and simple relationships to external stimuli or internal states. Our ability to record the detailed activity of single neurons, however, has led to a bias in searching for precisely these types of neurons. Recent advances in recording from larger populations of neurons in an unbiased manner reveal that complex response properties seem to be the rule, and simple neurons the exception. Using theoretical tools from the fields of Mathematics and Physics, along with analysis of experimental data from collaborating laboratories, my lab aims to understand the nature of neural representations.
The Synaptic Plasticity Laboratory of Senior Lecturer Shai Berlin. Study of synaptic plasticity using novel Optogenetics tools. We are interested in the molecular mechanisms governing synaptic plasticity and memory formation. How is information stored in neurons, in particular in dendritic spines? Spines are small, mushroom-like protrusions on dendrites that encompass the excitatory synapse, as well as a hundred more molecules and proteins. Biochemical and structural changes in the spine are thought to mediate ‘information’ storage. What are the changes that enable a spine to remember? How long can these changes last? Can these be modulated/controlled? To answer these questions, we employ a variety ofmethods ranging from molecular biology through electrophysiology and advanced imaging methods to optogenetics, to name a few. In particular, we design our own tools (optical and/or genetic) to monitor and control cellular processes to better understand synaptic plasticity.
 The Spatial Perception and Memory Laboratory of Associate Professor Dori Derdikman. How is space represented in the brain? How do we form our spatial memory? How are space and time related in the brain? What is the neuronal code for space? The Derdikman lab aims at researching these and related questions. We use diverse methods to conduct neuroscience research, including state-of-the-art electrophysiological recordings using tetrodes from awake-behaving animals, both tethered and telemetry-based, virtual-reality setups for animals, optogenetic methods, and mathematical modeling. The multi-disciplinary work in our lab hosts collaborations between biologists, electrical engineers, medical researchers, physicists, computer scientists, and mathematicians, all under the umbrella of brain science. We record neurons in the brain’s spatial cognitive map system, including grid cells in the entorhinal cortex, place cells in the hippocampus, and head-direction cells. We strive to understand the neural networks that encode our perception and dictate the animal’s behavior in such spaces.
The Spatial Perception and Memory Laboratory of Associate Professor Dori Derdikman. How is space represented in the brain? How do we form our spatial memory? How are space and time related in the brain? What is the neuronal code for space? The Derdikman lab aims at researching these and related questions. We use diverse methods to conduct neuroscience research, including state-of-the-art electrophysiological recordings using tetrodes from awake-behaving animals, both tethered and telemetry-based, virtual-reality setups for animals, optogenetic methods, and mathematical modeling. The multi-disciplinary work in our lab hosts collaborations between biologists, electrical engineers, medical researchers, physicists, computer scientists, and mathematicians, all under the umbrella of brain science. We record neurons in the brain’s spatial cognitive map system, including grid cells in the entorhinal cortex, place cells in the hippocampus, and head-direction cells. We strive to understand the neural networks that encode our perception and dictate the animal’s behavior in such spaces.
The Brain Systems Organization in Health and Disease Laboratory of Associate Professor Itamar Kahn. In my lab we seek to understand how systems-level dynamics give rise to various aspects of brain function and behavior in health and disease. To advance these goals, we take advantage of whole-brain functional imaging in humans and animal models. Functional magnetic resonance imaging (fMRI) allows us to measure activity in multiple brain systems simultaneously, and to look at dynamic interactions between regions of the brain. To manipulate well defined populations of neurons, we are using optogenetic techniques that enable cell-type specific optical control of electrical activity at a millisecond resolution. Combining optogenetic techniques with whole-brain fMRI (termed opto-fMRI), we study the mechanisms governing neural dynamics at the level of the microcircuit and across brain
regions.
 The Cortical Computation Lab of Professor Jackie Schiller. In my lab we investigate the fundamental mechanisms of information processing in dendrites of cortical neurons, and their possible contribution to the network function. In addition, we study the cellular mechanisms of memory and learning. Sensitive optical and electrophysiological tools are used to visualize and monitor neuronal function in the intact tissue. The lab is currently seeking a highly motivated graduate student or post-doc to lead projects dealing with questions related to the cellular basis of sensory processing and plasticity using combined microscopy, electrophysiology, histology, optogenetics and modeling methods both in-vitro in the slice preparation and in-vivo combining behavioral paradigms. For more details call 04-8295270 or send an e-mail to jackie@technion.ac.il.
The Cortical Computation Lab of Professor Jackie Schiller. In my lab we investigate the fundamental mechanisms of information processing in dendrites of cortical neurons, and their possible contribution to the network function. In addition, we study the cellular mechanisms of memory and learning. Sensitive optical and electrophysiological tools are used to visualize and monitor neuronal function in the intact tissue. The lab is currently seeking a highly motivated graduate student or post-doc to lead projects dealing with questions related to the cellular basis of sensory processing and plasticity using combined microscopy, electrophysiology, histology, optogenetics and modeling methods both in-vitro in the slice preparation and in-vivo combining behavioral paradigms. For more details call 04-8295270 or send an e-mail to jackie@technion.ac.il.
 The Epliepsy Research Lab of Associate Professor Yitzhak Schiller. The laboratory’s main research interests are neural network computations, information processing in the motor cortex and basal ganglia, mechanisms underlying ictogenesis in epilepsy, and development of novel neurostimulators and chemo-genetic based treatments for intractable epilepsy. The lab is seeking highly motivated graduate students to lead projects aimed at understanding the pathophysiology of the motor system (in particular in Parkinson’s disease) and epilepsy as well as test novel therapeutic approaches. For more details call 04-8295270 or send an e-mail to syitzhak@technion.ac.il.
The Epliepsy Research Lab of Associate Professor Yitzhak Schiller. The laboratory’s main research interests are neural network computations, information processing in the motor cortex and basal ganglia, mechanisms underlying ictogenesis in epilepsy, and development of novel neurostimulators and chemo-genetic based treatments for intractable epilepsy. The lab is seeking highly motivated graduate students to lead projects aimed at understanding the pathophysiology of the motor system (in particular in Parkinson’s disease) and epilepsy as well as test novel therapeutic approaches. For more details call 04-8295270 or send an e-mail to syitzhak@technion.ac.il.
 The Vascular Biology Laboratory of Associate Professor Zaid Abassi. Mechanisms of sodium/water retention and cardiac hypertrophy in congestive heart failure. Involvement of the endothelin and nitric oxide systems in the pathogenesis of cardiovascular and metabolic diseases. Pathogenesis of proteinuria in experimental models of nephrotic syndrome: Novel therapeutic approaches. Adverse renal effects of pneumoperitoneum: Mechanisms and therapeutic approaches. Acute kidney injury-Pathogenesis, detection and therapy.
The Vascular Biology Laboratory of Associate Professor Zaid Abassi. Mechanisms of sodium/water retention and cardiac hypertrophy in congestive heart failure. Involvement of the endothelin and nitric oxide systems in the pathogenesis of cardiovascular and metabolic diseases. Pathogenesis of proteinuria in experimental models of nephrotic syndrome: Novel therapeutic approaches. Adverse renal effects of pneumoperitoneum: Mechanisms and therapeutic approaches. Acute kidney injury-Pathogenesis, detection and therapy.
The Genome Structure and Function Laboratoryof Assistant Professor Noam Kaplan studies the profound connection between genetic information and its physical organization. We use a combination of advanced computational and experimental methods to decipher how the genome encodes its 3D organization and how this mediates biological function across different biological systems and in disease.
Biophysics & Systems Biology Laboratory of Assistant Professor Yonatan Savir focuses on machineries by which cells sense their environment and accordingly regulate their growth and metabolism; the impacts of age on signal processing networks in cells; information processing in biological systems.
The laboratory of Professor Shimon Marom
Concrete Research Interest: Biophysical and functional aspects of bio-electricity in proteins, cells and networks, focusing on mechanisms underlying emergence, dynamics and adaptation of bioelectrical phenomena at extended timescales.
Broader Interest: Advancing a framework of relational physiology, inspired by old and more recent ideas of relationists in a range of fields, from the science of self-organization and adaptivity, through theoretical biology, to psychology and anthropology. Within the physiological context, a relational approach entails focusing on the functional organization of systems that are embedded in a responsive and adaptive environment, analyzing processes rather than objects, implementing natural input statistics and closed-loop experimental designs.
 The Emerging Parasites Research Laboratory of Associate Professor Serge Ankri. Infections by parasitic protozoans are largely neglected. With descriptions of the human microbiota, new frontiers of investigation are developing to decipher the complexity of host–parasite–bacteria relationships. Amebiasis, a worldwide intestinal parasitic disease, is due to Entamoeba histolytica. In endemic areas, mixed intestinal infections of E. histolyticaand enteropathogenic Escherichia coliare common. Our laboratory has shown that Escherichia coliO55, protects the parasite against oxidative stress (OS) and induces transcriptome and redox-proteome changes in E.histolytica. Aiming to provide new strategies against amebiasis, we are studying the contribution to OS resistance and proficient intestinal colonization ofE. histolyticaand bacterial factors identified by omics.
The Emerging Parasites Research Laboratory of Associate Professor Serge Ankri. Infections by parasitic protozoans are largely neglected. With descriptions of the human microbiota, new frontiers of investigation are developing to decipher the complexity of host–parasite–bacteria relationships. Amebiasis, a worldwide intestinal parasitic disease, is due to Entamoeba histolytica. In endemic areas, mixed intestinal infections of E. histolyticaand enteropathogenic Escherichia coliare common. Our laboratory has shown that Escherichia coliO55, protects the parasite against oxidative stress (OS) and induces transcriptome and redox-proteome changes in E.histolytica. Aiming to provide new strategies against amebiasis, we are studying the contribution to OS resistance and proficient intestinal colonization ofE. histolyticaand bacterial factors identified by omics.
 The Regulation of Gene Expression Lab of Professor Mordechai (Motti) Choder.Regulation of gene expression is the process that controls life and determines our phenotype in health and disease. Understanding this process is therefore the focus of research of thousands of groups across the world. Gene expression is traditionally viewed as a linear process divided into distinct stages (e.g., transcription, translation). We have shown that this view is oversimplified. Using the state of the art technologies, we found that all these stages communicate with each other. We are working on factors that shuttle between all these stages and integrate all of them into a system. Among other activities these factors also stimulate mRNA synthesis and degradation. Hence we proposed that gene expression is a circular process – a concept illustrated in the figure. Our work initiated a new direction in studying gene expression that highlight novel targets for controlling cell phenotype and functionality. See http://choder.net.technion.ac.il/research for further information.
The Regulation of Gene Expression Lab of Professor Mordechai (Motti) Choder.Regulation of gene expression is the process that controls life and determines our phenotype in health and disease. Understanding this process is therefore the focus of research of thousands of groups across the world. Gene expression is traditionally viewed as a linear process divided into distinct stages (e.g., transcription, translation). We have shown that this view is oversimplified. Using the state of the art technologies, we found that all these stages communicate with each other. We are working on factors that shuttle between all these stages and integrate all of them into a system. Among other activities these factors also stimulate mRNA synthesis and degradation. Hence we proposed that gene expression is a circular process – a concept illustrated in the figure. Our work initiated a new direction in studying gene expression that highlight novel targets for controlling cell phenotype and functionality. See http://choder.net.technion.ac.il/research for further information.
 The Molecular Virology Laboratory of Associate Professor Tamar Kleinberger. Research in the lab focuses on virus-host cell interactions and on the use of viral proteins for induction of cancer-specific cell death. Specifically, the lab currently studies the mechanisms involved in the ability of the adenovirus E4orf4 protein to inhibit the DNA damage response, a cellular network that identifies DNA damage and transmits signals that promote its repair. This system serves as an antiviral defense mechanism and must be neutralized to improve virus replication. The lab also investigates how E4orf4 and its cellular partners (in particular protein phosphatase 2A and chromatin factors) induce cell death. Understanding these mechanisms may provide an explanation for the cancer specificity of E4orf4-induced cell death and may, in the long run, suggest novel targets for cancer therapy.
The Molecular Virology Laboratory of Associate Professor Tamar Kleinberger. Research in the lab focuses on virus-host cell interactions and on the use of viral proteins for induction of cancer-specific cell death. Specifically, the lab currently studies the mechanisms involved in the ability of the adenovirus E4orf4 protein to inhibit the DNA damage response, a cellular network that identifies DNA damage and transmits signals that promote its repair. This system serves as an antiviral defense mechanism and must be neutralized to improve virus replication. The lab also investigates how E4orf4 and its cellular partners (in particular protein phosphatase 2A and chromatin factors) induce cell death. Understanding these mechanisms may provide an explanation for the cancer specificity of E4orf4-induced cell death and may, in the long run, suggest novel targets for cancer therapy.
 The Yeast Genetics Laboratory of Associate Professor Daniel Kornitzer. We investigate the pathogenic yeast Candida albicans, a prevalent cause of life-threatening infections in immunocompromised patients. We study the adaptations that turned this organism into a successful invader of patients’ blood and tissues, and focus, at the molecular level, on two of its pathogenic characteristics in particular. One is the ability of this fungus to utilize hemoglobin as an iron source, by relying on a newly identified family of extracellular heme-binding proteins. The second is its ability to switch from the yeast morphology to the more invasive mold morphology, and how this switch can be inhibited, i.a.via the activity of the ubiquitin system.
The Yeast Genetics Laboratory of Associate Professor Daniel Kornitzer. We investigate the pathogenic yeast Candida albicans, a prevalent cause of life-threatening infections in immunocompromised patients. We study the adaptations that turned this organism into a successful invader of patients’ blood and tissues, and focus, at the molecular level, on two of its pathogenic characteristics in particular. One is the ability of this fungus to utilize hemoglobin as an iron source, by relying on a newly identified family of extracellular heme-binding proteins. The second is its ability to switch from the yeast morphology to the more invasive mold morphology, and how this switch can be inhibited, i.a.via the activity of the ubiquitin system.
 The Developmental Immunology and Aging Laboratory of Professor Doron Melamed. which focuses on studying mechanisms controlling generation of normal B-lymphocytes and B cell malignancies, to understand how cellular pathways that include signaling molecules, transcription factors and microRNAs operate during B cell development and control crucial fate decisions of normal and transformed B cells. The lab is specifically interested in finding how sensitivity and resistance to cell death signals are regulated in these cells. An additional topic of interest is in investigating how homeostatic mechanisms control B lymphopiesis in general, and particularly in aging, though attempts to identify cross-talk mediators between peripheral B cells and progenitors in the bone marrow that can be targeted to restore immune competence in aging.
The Developmental Immunology and Aging Laboratory of Professor Doron Melamed. which focuses on studying mechanisms controlling generation of normal B-lymphocytes and B cell malignancies, to understand how cellular pathways that include signaling molecules, transcription factors and microRNAs operate during B cell development and control crucial fate decisions of normal and transformed B cells. The lab is specifically interested in finding how sensitivity and resistance to cell death signals are regulated in these cells. An additional topic of interest is in investigating how homeostatic mechanisms control B lymphopiesis in general, and particularly in aging, though attempts to identify cross-talk mediators between peripheral B cells and progenitors in the bone marrow that can be targeted to restore immune competence in aging.
 The Sleep and Psychoneuroimmunology Laboratory of Associate Professor Asya Rolls. Research in the lab focuses on the interaction between the emotional state and physical well-being. Using state of the art tools in neuroscience and in immunology the lab studies the neuronal basis of brain-immune communication, and in particular on two major aspects of brain activity; first, how does sleep and sleep disorders affect the immune system and second, how neuronal networks involved in positive emotions alter the ability of the immune system to fight infections and cancer.
The Sleep and Psychoneuroimmunology Laboratory of Associate Professor Asya Rolls. Research in the lab focuses on the interaction between the emotional state and physical well-being. Using state of the art tools in neuroscience and in immunology the lab studies the neuronal basis of brain-immune communication, and in particular on two major aspects of brain activity; first, how does sleep and sleep disorders affect the immune system and second, how neuronal networks involved in positive emotions alter the ability of the immune system to fight infections and cancer.
 Systems Immunology and Precision Medicine Lab of Assistant Professor Shai Shen-Orr. The immune system is at the heart of the majority of diseases. Yet, albeit extreme disease states, our current understanding of how the immune system varies over time and between individuals is close to nil. We hypothesize that the variability we observe among individuals forms a complex continuum of system states whose constraints we still do not understand, nor how it is affected by external (environmental) or internal (genetics) variability. To chart the immune system landscape and understand the forces shaping it, we are undertaking holistic studies, in which we comprehensively measure the immune system, predominantly its adaptation to chronic conditions. Specifically, in immune aging, inflammatory bowel disease and the inter-cellular interaction network governing cellular communication, including with the brain. These studies both inform our basic understanding of how emergent proprties of immunity arise as well as deliver actionable information to better precision medicine. Students in the lab come from diverse backgrounds, including biology, biotechnology, computer science and physics.
Systems Immunology and Precision Medicine Lab of Assistant Professor Shai Shen-Orr. The immune system is at the heart of the majority of diseases. Yet, albeit extreme disease states, our current understanding of how the immune system varies over time and between individuals is close to nil. We hypothesize that the variability we observe among individuals forms a complex continuum of system states whose constraints we still do not understand, nor how it is affected by external (environmental) or internal (genetics) variability. To chart the immune system landscape and understand the forces shaping it, we are undertaking holistic studies, in which we comprehensively measure the immune system, predominantly its adaptation to chronic conditions. Specifically, in immune aging, inflammatory bowel disease and the inter-cellular interaction network governing cellular communication, including with the brain. These studies both inform our basic understanding of how emergent proprties of immunity arise as well as deliver actionable information to better precision medicine. Students in the lab come from diverse backgrounds, including biology, biotechnology, computer science and physics.
 The Immune Cell Signaling, Migration and Cancer Laboratory of Assistant Professor Debbie Yablonski. Immune cells are remarkable for their extraordinary sensitivity to antigen, coupled with extreme selectivity. The lab studies the molecular regulatory mechanisms that underly this behavior, to explain how immune cells are able to mount potent responses to minute quantities of foreign antigens, while ignoring related “self” antigens. This question is studied in T cells, the central arm of the cellular immune system, and in mast cells, an important mediator of allergy. Within the cells, tightly regulated signal processing mechanisms are employed to integrate and interpret information about the presence of antigens, resulting in a cellular decision about how to respond. Specialized signaling proteins, known as adaptors, play an essential role in this process, and are the focus of the lab’s research. A variety of genetic approaches, including mouse models and somatic cell genome editing techniques, are used by the lab to explore the ways in which adaptor proteins fine tune immune sensitivity and selectivity.
The Immune Cell Signaling, Migration and Cancer Laboratory of Assistant Professor Debbie Yablonski. Immune cells are remarkable for their extraordinary sensitivity to antigen, coupled with extreme selectivity. The lab studies the molecular regulatory mechanisms that underly this behavior, to explain how immune cells are able to mount potent responses to minute quantities of foreign antigens, while ignoring related “self” antigens. This question is studied in T cells, the central arm of the cellular immune system, and in mast cells, an important mediator of allergy. Within the cells, tightly regulated signal processing mechanisms are employed to integrate and interpret information about the presence of antigens, resulting in a cellular decision about how to respond. Specialized signaling proteins, known as adaptors, play an essential role in this process, and are the focus of the lab’s research. A variety of genetic approaches, including mouse models and somatic cell genome editing techniques, are used by the lab to explore the ways in which adaptor proteins fine tune immune sensitivity and selectivity.
The Psychobiology Research Laboratory of Lecturer Alon Shamir, Mazor Mental health center – Acre. The laboratory of Dr. Shamir focuses on elucidating the behavioral, biochemical and molecular consequences of the NRG/ErbB signaling pathway in complex behaviors related to schizophrenia and other psychiatric disorder, on underlying the therapeutics potential of the pathway, and on the identification of biomarkers for schizophrenia.
Infectious Diseases Institute at the Rambam Health Care Campus of Associate Professor Michal Paul. Clinical / epidemiological studies in infectious diseases, multidrug-resistant bacteria, antibiotic therapy, systematic review/ meta-analyses in infectious diseases.
The Laboratory of Stem Cells Utilization in Diabetic Wound Healing of Assistant Professor Mogher Khamaisi, Rambam Health Care Campus. Research in our laboratory focuses on molecular and cellular mechanisms underlying impaired wound healing in diabetic patients, as well as evaluating the efficacy of patient derived induced pluripotent stem cells (iPSCs) in enhancing diabetic wound healing.
Laboratory for Bone Repair, CRIR of Senior Lecturer Hadar Zigdon Giladi, Rambam Health Care Campus. Research focus – study methods to induce bone regeneration using scaffolds, cells and growth factors. Specifically, we study the ability of endothelial progenitor cells (EPCs) to enhance bone regeneration.
Gynecology Research Laboratory of Associate Professor Moti HalakatHillel Yaffe Medical center.
The laboratory of Clinical Neurophysiology of Prof. David Yarnitsky explores pain processing in humans, in health and in disease. We employ tools such as psychophysical testing, electroencephalography (EEG) and event related potentials (ERP) as well as MRI imaging and psychological profiling in order to understand how pain processing done in health, how is it changed in disease, and how can this understanding help in tailoring individual pain treatment.